For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VQRRQAYVQAIHRAAAPGAGLFILAFAQRPFGDGMPGPNGFTAEELRDTVATQWTVDEVRPATLYANDVQ LAAGSAPGVEAQRDDKGRLMFPGFLLLAHKG
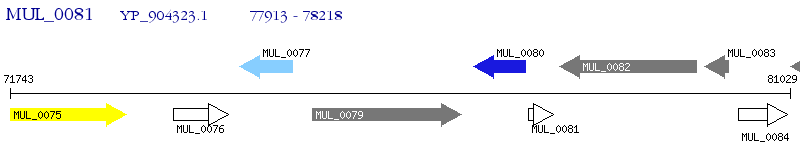
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0081 | - | - | 100% (101) | hypothetical protein MUL_0081 |
| M. ulcerans Agy99 | MUL_0662 | - | 7e-12 | 43.90% (82) | methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3725 | - | 2e-17 | 40.95% (105) | hypothetical protein Mb3725 |
| M. gilvum PYR-GCK | Mflv_1324 | - | 1e-18 | 42.57% (101) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv3699 | - | 3e-17 | 40.95% (105) | hypothetical protein Rv3699 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0961c | - | 3e-16 | 42.57% (101) | hypothetical protein MAB_0961c |
| M. marinum M | MMAR_0092 | - | 3e-53 | 98.02% (101) | hypothetical protein MMAR_0092 |
| M. avium 104 | MAV_0313 | - | 1e-22 | 49.02% (102) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. smegmatis MC2 155 | MSMEG_6235 | - | 4e-20 | 50.00% (102) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. thermoresistible (build 8) | TH_3824 | - | 2e-31 | 65.00% (100) | PUTATIVE thiopurine S-methyltransferase (tpmt) superfamily |
| M. vanbaalenii PYR-1 | Mvan_5471 | - | 7e-19 | 45.54% (101) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3725|M.bovis_AF2122/97 ---MTDEVMDWDSAYREQGA-FEGPPPWNIGEPQPELATLIAA-------
Rv3699|M.tuberculosis_H37Rv ---MTDEVMDWDSAYREQGA-FEGPPPWNIGEPQPELATLIAA-------
MSMEG_6235|M.smegmatis_MC2_155 MGVMTSHVLDWDGAYRGDTG-FQGPPPWNIGEPQPELAALHRA-------
Mflv_1324|M.gilvum_PYR-GCK ----MAEVMDWDSAYRGEGE-FEGEPPWNIGEPQPELAALWRD-------
Mvan_5471|M.vanbaalenii_PYR-1 -MAHMTDVMDWDSAYRGEGD-FEGEPPWNIGEPQPELAALWRD-------
MAB_0961c|M.abscessus_ATCC_199 --MTSNELPDWDATYQGKGELFQGEPPWNIGEPQPELAALIDA-------
MUL_0081|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0092|M.marinum_M --------------------------------------------------
TH_3824|M.thermoresistible__bu --------MEWDDMYRRGVP-----PPWSIGAPQPELAALLDAGPAQCPV
MAV_0313|M.avium_104 ---MTQPDTDWDAAYRQAAP-----PPWSIGRPQPELERLIDE-------
Mb3725|M.bovis_AF2122/97 -GKVRSDVLDAGCGYAELSLALAADGYTVVGIDLTPTAVAAATKAAEERG
Rv3699|M.tuberculosis_H37Rv -GKVRSDVLDAGCGYAELSLALAADGYTVVGIDLTPTAVAAATKAAEERG
MSMEG_6235|M.smegmatis_MC2_155 -GRFRSDVLDAGCGHAELSLALAADGYTVVGMDLSPSAIAAANRAAQERN
Mflv_1324|M.gilvum_PYR-GCK -GRFRSEVLDAGCGHAELSLALAADGYTVTGVDISPTAMAAASAAAAERG
Mvan_5471|M.vanbaalenii_PYR-1 -GKFRSEVLDAGCGHAELSLALAADGYTVTGVDLSPTAVAAATAASAERG
MAB_0961c|M.abscessus_ATCC_199 -GKFHGTVLDVGCGHAETSLRLAALGHTTVGLDLSPTAIEAARAAAAERG
MUL_0081|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0092|M.marinum_M --------------------------------------------------
TH_3824|M.thermoresistible__bu QGTVRGEVLDAGCGEAALSLELAARGHTVVGLDCSAAAVAAATATAAERG
MAV_0313|M.avium_104 -GKFRSDVLDSGCGHAALSLRLAALGHTVVGLDASATAIAEATAAAAAQG
Mb3725|M.bovis_AF2122/97 LTT-ASFVQADITEFAAYPAGSAGRFSTVIDSTLFHSLPVDSRDRYLSSV
Rv3699|M.tuberculosis_H37Rv LTT-ASFVQADITEFAAYPAGSAGRFSTVIDSTLFHSLPVDSRDRYLSSV
MSMEG_6235|M.smegmatis_MC2_155 LAT-ASFVQGDITSFTGY----DGRFSTVIDSTLFHSLPVEGREGYLQSI
Mflv_1324|M.gilvum_PYR-GCK LSEKASFVAADITALSGF----DGRFSTVVDSTLFHSLPVESRAGYLESV
Mvan_5471|M.vanbaalenii_PYR-1 LSERTSFVAADITTLTGF----DGRFATVVDSTLFHSLPVESRDGYLQSV
MAB_0961c|M.abscessus_ATCC_199 LTN-ATFEVADITDFSGY----DGRFNTIVDSTLFHSIPVEAREAYLQSI
MUL_0081|M.ulcerans_Agy99 ---------------------------------------MQRRQAYVQAI
MMAR_0092|M.marinum_M ---------------------------------------MQRRQAYVQAI
TH_3824|M.thermoresistible__bu LTT-ATFAQADLTEFGGY----DGRFSTIIDSGLLHALPVDRRQAYLRAI
MAV_0313|M.avium_104 LTT-ATFARADVTDFADYPPGSEGRFATIVDSGLFHALPPQRRQDYLRSI
: * *: ::
Mb3725|M.bovis_AF2122/97 HRAAAPGASYYVLVFAKGAFPAELEV--KPNEVDEDELRAAVSKYWKIDE
Rv3699|M.tuberculosis_H37Rv HRAAAPGASYYVLVFAKGAFPAELEV--KPNEVDEDELRAAVSKYWKIDE
MSMEG_6235|M.smegmatis_MC2_155 HRASAPGARYFVLVFAKGAFPADLEV--KPNEVTEEELRTTVGKYWVVDD
Mflv_1324|M.gilvum_PYR-GCK HRAARPGAGFFVLVFAKGAFPADTEQ--GPNEVSEIELREAVSPYWTIDD
Mvan_5471|M.vanbaalenii_PYR-1 HRAAAPGAGYFVLVFAKGAFPAEMEG--GPNEVTELELREAVSRYWIIDD
MAB_0961c|M.abscessus_ATCC_199 SRAAAPDASYFVLVFAVGAFPPGIG----PNAVTEDELRAAVEPYWAIDE
MUL_0081|M.ulcerans_Agy99 HRAAAPGAGLFILAFAQRPFGDGMP---GPNGFTAEELRDTVATQWTVDE
MMAR_0092|M.marinum_M HRAAAPGAGLFILAFAQRQFGDGMP---GPNGFTAEELRDTVATQWTVDE
TH_3824|M.thermoresistible__bu HRAAAPNARLFILTFAERPFGDGH----GPDGFTAEELRDTVGVLWTVDE
MAV_0313|M.avium_104 FRAAAPGAALYILAFAAGALAPAHPDRPGPQGFTETELREAVSVLWHIDD
**: *.* ::*.** : *: . *** :* * :*:
Mb3725|M.bovis_AF2122/97 IRPAFIHVNPVTIPPQLAGAPVEFPPYDHDEKGRVKFPAYLLTAHKAG
Rv3699|M.tuberculosis_H37Rv IRPAFIHVNPVTIPPQLAGAPVEFPPYDHDEKGRVKFPAYLLTAHKAG
MSMEG_6235|M.smegmatis_MC2_155 IRPAFIHANAVVFP----DAP-ELPPHGYDDKGRIMFPAFLLEAHRD-
Mflv_1324|M.gilvum_PYR-GCK IRPALIHTNVPKLP----GMP-PPPLDMLDDRGHLRMQAFLLSAHKEP
Mvan_5471|M.vanbaalenii_PYR-1 IRPALIDTNVPKLP----GMP-PPPLDMLDDRGRLRMQAFLLQAHKES
MAB_0961c|M.abscessus_ATCC_199 LSSAYIHSNIPEVIP---GANFEMPVFHKDEKGRNKQPAFLLQAHKR-
MUL_0081|M.ulcerans_Agy99 VRPATLYANDVQLA----AGSAPGVEAQRDDKGRLMFPGFLLLAHKG-
MMAR_0092|M.marinum_M VRSATLYANDVQLA----AGSAPGVEAQRDDKGRLMFPGFLLLAHKG-
TH_3824|M.thermoresistible__bu IRPAKLYVNDFTGD----LLDGPPTAYERAADGKLMFPGFLVSAHKD-
MAV_0313|M.avium_104 LRAARVYGNDDSAG----APDSPLAHLERDGEGHFMAPGFLVSAHKRD
: .* : * *: .:*: **: