For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPTALITGASGAIGSAIAAALAPTHTLLLAGRPSARLDEVAERLGAPTWPLDLTDADSIESSTEVLAELD VLVHNAGVLFPGRVGESTADEWRASFEVNVTGAVALTLALLPALRAARGHVVFINSGAGRKVSAGMASYS ASKFALRAFADSLRADEPSLRVTSVFPGRTDSDMQRELIAYEGREYDPGSFLRPETVAGLVASAVNTPRD GHVHEIVVRPG
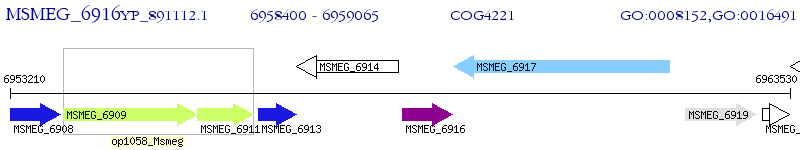
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6916 | - | - | 100% (221) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0258 | - | 4e-23 | 33.33% (225) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. smegmatis MC2 155 | MSMEG_0930 | - | 1e-20 | 31.90% (232) | serine 3-dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0788c | - | 7e-21 | 29.46% (241) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0854 | - | 7e-87 | 70.00% (220) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv0765c | - | 7e-21 | 29.46% (241) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01094 | - | 2e-15 | 31.28% (195) | short chain alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4922 | - | 3e-72 | 61.64% (219) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_0058 | - | 2e-90 | 73.64% (220) | short-chain type oxidoreductase |
| M. avium 104 | MAV_0056 | - | 2e-89 | 75.00% (220) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_0508 | - | 4e-83 | 67.27% (220) | short chain dehydrogenase |
| M. ulcerans Agy99 | MUL_0057 | - | 5e-89 | 73.18% (220) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_6051 | - | 1e-87 | 70.91% (220) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0788c|M.bovis_AF2122/97 MPRFEPHPARRTTVVAGASSGIGAATATELAGRGFPVALGARRMDKLAEL
Rv0765c|M.tuberculosis_H37Rv MPRFEPHPARRTTVVAGASSGIGAATATELAGRGFPVALGARRMDKLAEL
MMAR_0058|M.marinum_M MP---------TALITGASRGIGAAIAAALAPT-HTLLLAGRPSSQLDAV
MUL_0057|M.ulcerans_Agy99 MP---------TALITGASRGIGAAIAAALAPT-HTLLLAGRPSAQLDAV
MAV_0056|M.avium_104 MP---------NALITGAGGGIGSAIATALAPT-HTLLLAGRPSDRLDAV
MSMEG_6916|M.smegmatis_MC2_155 MP---------TALITGASGAIGSAIAAALAPT-HTLLLAGRPSARLDEV
Mflv_0854|M.gilvum_PYR-GCK MP---------TAMITGASGGLGSALADALAPT-HTLFLAGRPSPRLDAV
Mvan_6051|M.vanbaalenii_PYR-1 MP---------TAMITGASGGVGSALADALAPT-HTLFLAGRPSPRLDAV
MAB_4922|M.abscessus_ATCC_1997 MP---------HALITGAAGGLGAAIARALAAT-HSLLLGGRPSLRLDTL
TH_0508|M.thermoresistible__bu MP---------TAMITGAAGGLGSALTAALAPT-HTLFLAGRASDRLDDL
MLBr_01094|M.leprae_Br4923 MEGFAG----KVAVVTGAGSGIGQALAIELARSGAKLAISDVDGEGLAQT
* ::::**. .:* * : ** : :. *
Mb0788c|M.bovis_AF2122/97 VDKIRADGGEAVAFPLDVTDPESVKSFVAQTVEALGEVELLVSSAGDMLP
Rv0765c|M.tuberculosis_H37Rv VDKIRADGGEAVAFPLDVTDPESVKSFVAQTVEALGEVELLVSSAGDMLP
MMAR_0058|M.marinum_M ADQFGAT-----TFPLDLTDDE----SIETSCEIVDELDVLIHNAGLSIP
MUL_0057|M.ulcerans_Agy99 ADQFGAT-----TFPLDLTDDE----SIETSCEIVDELDVLIHNAGLSIP
MAV_0056|M.avium_104 AQRLGAT-----TFPLDLTDGG----DIEAACEVVDELDVLVHNAGLSIP
MSMEG_6916|M.smegmatis_MC2_155 AERLGAP-----TWPLDLTDAD----SIESSTEVLAELDVLVHNAGVLFP
Mflv_0854|M.gilvum_PYR-GCK AERLGAT-----TWPVDLADPD----AMAAVVEPIVELDVLIHNAGVAYP
Mvan_6051|M.vanbaalenii_PYR-1 AERLGAT-----TWPVDLADPD----AIPAVVEPIVELDVLIHNAGVAYP
MAB_4922|M.abscessus_ATCC_1997 ADELDAT-----PWPVDLSDQD----AILAAVAGIDALDVLVHNAGVSYP
TH_0508|M.thermoresistible__bu AQRYGAT-----TWPVDFTDLE----ALPAVVEPIDELDVLIHNAGVTHP
MLBr_01094|M.leprae_Br4923 EGQLKAIGASARTDRLDVTEREAFLTYADVVHENFGKVNQIYNNAGIAFT
. * . :*.:: . :: : .** .
Mb0788c|M.bovis_AF2122/97 GQLHEVSTEAFAEQVQIHLVGANRLATAVLPAMVARRRGDLIFVGSDVGL
Rv0765c|M.tuberculosis_H37Rv GQLHEVSTEAFAEQVQIHLVGANRLATAVLPAMVARRRGDLIFVGSDVGL
MMAR_0058|M.marinum_M GHFGDSHVDEWRATFNVNLFGAVALTLALLPALRSAR-GQVVFINSGAGR
MUL_0057|M.ulcerans_Agy99 GHFGDSHVDEWRATFNVNLFGAVALTLALLPAPRSAR-GQVVFINSGAGR
MAV_0056|M.avium_104 GNVADSNVDEWRATFAVNVFGPVELTLALLPALRRAR-GQVVFINSGAGR
MSMEG_6916|M.smegmatis_MC2_155 GRVGESTADEWRASFEVNVTGAVALTLALLPALRAAR-GHVVFINSGAGR
Mflv_0854|M.gilvum_PYR-GCK GRVAESHVDEWRSTMAVNVVGAVALTLELLPALRAAR-GHVVFINSGAGI
Mvan_6051|M.vanbaalenii_PYR-1 GRVAESDVDQWRATMAVNVVGAVALTLELLPALRSAG-GHVVFINSGAGI
MAB_4922|M.abscessus_ATCC_1997 ATIADSTLADWRKTLDVNVLAPVALTQALLPALRAAQ-GDVVFINSGAGI
TH_0508|M.thermoresistible__bu ADFADSTVEEWRNSFDVNVIGPVALTQALLPALRQRR-GHVVIINSGSGL
MLBr_01094|M.leprae_Br4923 GDVEVSHFKDIERVMDVDYWGVVNGTKAFLPYLISSGDGHVINISSVFGL
. . . :. . : .** *.:: :.* *
Mb0788c|M.bovis_AF2122/97 RQRPHMGAYGAAKAGLAAMVTNLQMELEGT--GVRASIVHPGPTLTGMGW
Rv0765c|M.tuberculosis_H37Rv RQRPHMGAYGAAKAGLAAMVTNLQMELEGT--GVRASIVHPGPTLTGMGW
MMAR_0058|M.marinum_M NVSAGMASYSASKFALRAFADSLRTDEP----TLRVTTVYPGRTDTDMQR
MUL_0057|M.ulcerans_Agy99 NVSAGMASYSASKFALRAFADSLRTDES----TLRVTTVYPGRTDTDMQR
MAV_0056|M.avium_104 NVSPGMASYSASKFALRAFADSLRNDEP----ELRVTTVYPGRTDTGMQR
MSMEG_6916|M.smegmatis_MC2_155 KVSAGMASYSASKFALRAFADSLRADEP----SLRVTSVFPGRTDSDMQR
Mflv_0854|M.gilvum_PYR-GCK NASPGLASYTASKFALRGFADSLRADEP----TLRVTSVHPGRIATAMQE
Mvan_6051|M.vanbaalenii_PYR-1 NASPGLASYSASKFALRAFADSLRADEP----GLRVTSVHPGRIATAMQE
MAB_4922|M.abscessus_ATCC_1997 NAHPGIGSYSVSKFALRGFADVLRAEEP----ALRVTSVHPGRISTPMQQ
TH_0508|M.thermoresistible__bu RAHAGMASYSASKFAARAFADALREEEP----LLRVTSVFPGRIDTAMQQ
MLBr_01094|M.leprae_Br4923 FSVPGQAAYNSAKFAVRGFTEALREEMALAGRPVNVTTVYPGGIKTAIAR
. .:* :* . .:. *: : :..: *.** : :
Mb0788c|M.bovis_AF2122/97 QLSAEQVGPMLADWAKWGQARHNYFLRPSDLARAIAFVAETP-------R
Rv0765c|M.tuberculosis_H37Rv QLSAEQVGPMLADWAKWGQARHNYFLRPSDLARAIAFVAETP-------R
MMAR_0058|M.marinum_M ELVDFEG----------GTYDPARFMRADTVAAVVANVVATP-------P
MUL_0057|M.ulcerans_Agy99 ELVDFEG----------GTYDPARFMRADTVAEVVANVVATP-------P
MAV_0056|M.avium_104 ELIAFEG----------GSYDPDRFLKPETVAAAVANVVATP-------P
MSMEG_6916|M.smegmatis_MC2_155 ELIAYEG----------REYDPGSFLRPETVAGLVASAVNTP-------R
Mflv_0854|M.gilvum_PYR-GCK DLVAYEG----------RPYEPDRFLSPQTVARLTVDAINAP-------A
Mvan_6051|M.vanbaalenii_PYR-1 DLVAYEG----------RPYDPAQFLSPQTVAQVTADAINAP-------A
MAB_4922|M.abscessus_ATCC_1997 ELTEHEG----------REYNPDDYMRPESVAQLVADAVNLP-------R
TH_0508|M.thermoresistible__bu KLVAYEG----------RQYNPDEFLRPETVAKIVADAVNTP-------P
MLBr_01094|M.leprae_Br4923 NATAAEG--LDVSKIASRFDTWVAHTSPQHAARIILKAVRKKKARVLVGP
. : . .. * .
Mb0788c|M.bovis_AF2122/97 GCVVVNMEIQPEAPLRDAPAHRQKLVLGEEGMPG
Rv0765c|M.tuberculosis_H37Rv GCVVVNMEIQPEAPLRDAPAHRQKLVLGEEGMPG
MMAR_0058|M.marinum_M DGHVHEVVIRPR----------------------
MUL_0057|M.ulcerans_Agy99 DGHVHEVVIRPR----------------------
MAV_0056|M.avium_104 DGHVHEVVLRPARR--------------------
MSMEG_6916|M.smegmatis_MC2_155 DGHVHEIVVRPG----------------------
Mflv_0854|M.gilvum_PYR-GCK DAHVHEVIIRPSPAPR------------------
Mvan_6051|M.vanbaalenii_PYR-1 DAHIHEVIVRPR----------------------
MAB_4922|M.abscessus_ATCC_1997 DARVHQIVVRQN----------------------
TH_0508|M.thermoresistible__bu DAHLHEIVVRPYQ---------------------
MLBr_01094|M.leprae_Br4923 DAKVANVVVRFSGGAGYQRLFAQVASRLILNQR-
. : :: ::