For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPHALITGAAGGLGAAIARALAATHSLLLGGRPSLRLDTLADELDATPWPVDLSDQDAILAAVAGIDALD VLVHNAGVSYPATIADSTLADWRKTLDVNVLAPVALTQALLPALRAAQGDVVFINSGAGINAHPGIGSYS VSKFALRGFADVLRAEEPALRVTSVHPGRISTPMQQELTEHEGREYNPDDYMRPESVAQLVADAVNLPRD ARVHQIVVRQN
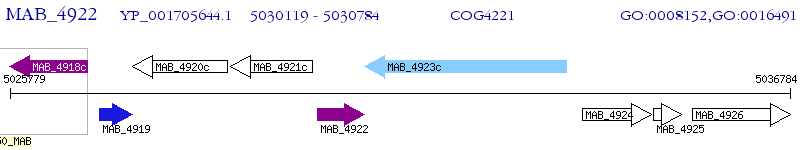
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4922 | - | - | 100% (221) | putative short-chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_4060 | - | 7e-23 | 30.84% (227) | putative short chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_3420c | - | 2e-20 | 33.70% (184) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0494c | - | 2e-20 | 31.58% (228) | putative short-chain type oxidoreductase |
| M. gilvum PYR-GCK | Mflv_0854 | - | 3e-78 | 63.01% (219) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv0484c | - | 1e-20 | 32.02% (228) | short-chain type oxidoreductase |
| M. leprae Br4923 | MLBr_01740 | - | 7e-18 | 34.78% (184) | short chain dehydrogenase |
| M. marinum M | MMAR_0058 | - | 8e-67 | 57.08% (219) | short-chain type oxidoreductase |
| M. avium 104 | MAV_0056 | - | 3e-70 | 60.73% (219) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6916 | - | 4e-72 | 61.64% (219) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_0508 | - | 8e-79 | 64.84% (219) | short chain dehydrogenase |
| M. ulcerans Agy99 | MUL_0057 | - | 1e-65 | 56.16% (219) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_6051 | - | 1e-76 | 62.56% (219) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0494c|M.bovis_AF2122/97 --MTTIGTRKRVAVVTGASSGIGEATARTLAAQGFHVVAVARRADRITAL
Rv0484c|M.tuberculosis_H37Rv --MTTIGTRKRVAVVTGASSGIGEATARTLAAQGFHVVAVARRADRITAL
Mflv_0854|M.gilvum_PYR-GCK ---------MPTAMITGASGGLGSALADALAPT-HTLFLAGRPSPRLDAV
Mvan_6051|M.vanbaalenii_PYR-1 ---------MPTAMITGASGGVGSALADALAPT-HTLFLAGRPSPRLDAV
MMAR_0058|M.marinum_M ---------MPTALITGASRGIGAAIAAALAPT-HTLLLAGRPSSQLDAV
MUL_0057|M.ulcerans_Agy99 ---------MPTALITGASRGIGAAIAAALAPT-HTLLLAGRPSAQLDAV
MAV_0056|M.avium_104 ---------MPNALITGAGGGIGSAIATALAPT-HTLLLAGRPSDRLDAV
MSMEG_6916|M.smegmatis_MC2_155 ---------MPTALITGASGAIGSAIAAALAPT-HTLLLAGRPSARLDEV
TH_0508|M.thermoresistible__bu ---------MPTAMITGAAGGLGSALTAALAPT-HTLFLAGRASDRLDDL
MAB_4922|M.abscessus_ATCC_1997 ---------MPHALITGAAGGLGAAIARALAAT-HSLLLGGRPSLRLDTL
MLBr_01740|M.leprae_Br4923 MVKTAQYFVGKRCFVTGAASGIGRATALRLAAYGAELYLTDRHCDGLAQT
..:***. .:* * : **. : * . :
Mb0494c|M.bovis_AF2122/97 ANQIGGT------AIVADVTDDAAVEALARALSR----VDVLVNNAGGAK
Rv0484c|M.tuberculosis_H37Rv ANQIGGT------AIVADVTDDAAVEALARALSR----VDVLVNNAGGAK
Mflv_0854|M.gilvum_PYR-GCK AERLGAT------TWPVDLADPDAMAAVVEPIVE----LDVLIHNAG-VA
Mvan_6051|M.vanbaalenii_PYR-1 AERLGAT------TWPVDLADPDAIPAVVEPIVE----LDVLIHNAG-VA
MMAR_0058|M.marinum_M ADQFGAT------TFPLDLTDDESIETSCEIVDE----LDVLIHNAG-LS
MUL_0057|M.ulcerans_Agy99 ADQFGAT------TFPLDLTDDESIETSCEIVDE----LDVLIHNAG-LS
MAV_0056|M.avium_104 AQRLGAT------TFPLDLTDGGDIEAACEVVDE----LDVLVHNAG-LS
MSMEG_6916|M.smegmatis_MC2_155 AERLGAP------TWPLDLTDADSIESSTEVLAE----LDVLVHNAG-VL
TH_0508|M.thermoresistible__bu AQRYGAT------TWPVDFTDLEALPAVVEPIDE----LDVLIHNAG-VT
MAB_4922|M.abscessus_ATCC_1997 ADELDAT------PWPVDLSDQDAILAAVAGIDA----LDVLVHNAG-VS
MLBr_01740|M.leprae_Br4923 VSEARALGAQVPEYRALDISDYDEVAAFGADIHARHPSMDIVMNIAG-VS
... . *.:* : : : :*:::: **
Mb0494c|M.bovis_AF2122/97 GLQFVADADLEHWRWMWDTNVLGTLRVTRALLPKLIDSGDG-LIVTVTSI
Rv0484c|M.tuberculosis_H37Rv GLQFVADADLEHWRWMWDTNVLGTLRVTRALLPKLIDSGDG-LIVTVTSI
Mflv_0854|M.gilvum_PYR-GCK YPGRVAESHVDEWRSTMAVNVVGAVALTLELLPALR-AARG-HVVFINSG
Mvan_6051|M.vanbaalenii_PYR-1 YPGRVAESDVDQWRATMAVNVVGAVALTLELLPALR-SAGG-HVVFINSG
MMAR_0058|M.marinum_M IPGHFGDSHVDEWRATFNVNLFGAVALTLALLPALR-SARG-QVVFINSG
MUL_0057|M.ulcerans_Agy99 IPGHFGDSHVDEWRATFNVNLFGAVALTLALLPAPR-SARG-QVVFINSG
MAV_0056|M.avium_104 IPGNVADSNVDEWRATFAVNVFGPVELTLALLPALR-RARG-QVVFINSG
MSMEG_6916|M.smegmatis_MC2_155 FPGRVGESTADEWRASFEVNVTGAVALTLALLPALR-AARG-HVVFINSG
TH_0508|M.thermoresistible__bu HPADFADSTVEEWRNSFDVNVIGPVALTQALLPALR-QRRG-HVVIINSG
MAB_4922|M.abscessus_ATCC_1997 YPATIADSTLADWRKTLDVNVLAPVALTQALLPALR-AAQG-DVVFINSG
MLBr_01740|M.leprae_Br4923 VWGTVDRLTHDQWSNVVAINLMGPIHIIETFVPAILAAGRGGHLVNVSSA
. .* *: ..: : ::* * :* :.*
Mb0494c|M.bovis_AF2122/97 AALEVYDGGAGYTAAKHAQGALHRTLRGELLGKPVRLTEIAPGAVETEFS
Rv0484c|M.tuberculosis_H37Rv AAIEVYDGGAGYTAAKHAQGALHRTLRGELLGKPVRLTEIAPGAVETEFS
Mflv_0854|M.gilvum_PYR-GCK AGINASPGLASYTASKFALRGFADSLRADEP--TLRVTSVHPGRIATAMQ
Mvan_6051|M.vanbaalenii_PYR-1 AGINASPGLASYSASKFALRAFADSLRADEP--GLRVTSVHPGRIATAMQ
MMAR_0058|M.marinum_M AGRNVSAGMASYSASKFALRAFADSLRTDEP--TLRVTTVYPGRTDTDMQ
MUL_0057|M.ulcerans_Agy99 AGRNVSAGMASYSASKFALRAFADSLRTDES--TLRVTTVYPGRTDTDMQ
MAV_0056|M.avium_104 AGRNVSPGMASYSASKFALRAFADSLRNDEP--ELRVTTVYPGRTDTGMQ
MSMEG_6916|M.smegmatis_MC2_155 AGRKVSAGMASYSASKFALRAFADSLRADEP--SLRVTSVFPGRTDSDMQ
TH_0508|M.thermoresistible__bu SGLRAHAGMASYSASKFAARAFADALREEEP--LLRVTSVFPGRIDTAMQ
MAB_4922|M.abscessus_ATCC_1997 AGINAHPGIGSYSVSKFALRGFADVLRAEEP--ALRVTSVHPGRISTPMQ
MLBr_01740|M.leprae_Br4923 AGLVALPWHAAYSASKYGLRGLSEVLRFDLARYRIGVSVVVPGAVRTPLV
:. . ..*:.:*.. .: ** : : :: : ** : :
Mb0494c|M.bovis_AF2122/97 --LVRFDGDQQ-----RADAVYAGMTPLVAADVAEVIGFVAT------RP
Rv0484c|M.tuberculosis_H37Rv --LVRFDGDQQ-----RADAVYAGMTPLVAADVAEVIGFVAT------RP
Mflv_0854|M.gilvum_PYR-GCK EDLVAYEGRP-----------YEPDRFLSPQTVARLTVDAIN------AP
Mvan_6051|M.vanbaalenii_PYR-1 EDLVAYEGRP-----------YDPAQFLSPQTVAQVTADAIN------AP
MMAR_0058|M.marinum_M RELVDFEGGT-----------YDPARFMRADTVAAVVANVVA------TP
MUL_0057|M.ulcerans_Agy99 RELVDFEGGT-----------YDPARFMRADTVAEVVANVVA------TP
MAV_0056|M.avium_104 RELIAFEGGS-----------YDPDRFLKPETVAAAVANVVA------TP
MSMEG_6916|M.smegmatis_MC2_155 RELIAYEGRE-----------YDPGSFLRPETVAGLVASAVN------TP
TH_0508|M.thermoresistible__bu QKLVAYEGRQ-----------YNPDEFLRPETVAKIVADAVN------TP
MAB_4922|M.abscessus_ATCC_1997 QELTEHEGRE-----------YNPDDYMRPESVAQLVADAVN------LP
MLBr_01740|M.leprae_Br4923 DTIEIAGVDRQDPRFSRWVDRFRGHAVSAERAAEKILAGVAKNRYLIYTS
: : . . .
Mb0494c|M.bovis_AF2122/97 SHV-------------------NLDQIVIRPRDQASASRRATHPVR----
Rv0484c|M.tuberculosis_H37Rv SHV-------------------NLDQIVIRPRDQASASRRATHPVR----
Mflv_0854|M.gilvum_PYR-GCK ADA-------------------HVHEVIIRPSPAPR--------------
Mvan_6051|M.vanbaalenii_PYR-1 ADA-------------------HIHEVIVRPR------------------
MMAR_0058|M.marinum_M PDG-------------------HVHEVVIRPR------------------
MUL_0057|M.ulcerans_Agy99 PDG-------------------HVHEVVIRPR------------------
MAV_0056|M.avium_104 PDG-------------------HVHEVVLRPARR----------------
MSMEG_6916|M.smegmatis_MC2_155 RDG-------------------HVHEIVVRPG------------------
TH_0508|M.thermoresistible__bu PDA-------------------HLHEIVVRPYQ-----------------
MAB_4922|M.abscessus_ATCC_1997 RDA-------------------RVHQIVVRQN------------------
MLBr_01740|M.leprae_Br4923 HDIRVLYGFKRLAWWPYGVVMKQVNVVFTRALRPSPATMRCVEPIKVGVY
. .:. :. *
Mb0494c|M.bovis_AF2122/97 -------------
Rv0484c|M.tuberculosis_H37Rv -------------
Mflv_0854|M.gilvum_PYR-GCK -------------
Mvan_6051|M.vanbaalenii_PYR-1 -------------
MMAR_0058|M.marinum_M -------------
MUL_0057|M.ulcerans_Agy99 -------------
MAV_0056|M.avium_104 -------------
MSMEG_6916|M.smegmatis_MC2_155 -------------
TH_0508|M.thermoresistible__bu -------------
MAB_4922|M.abscessus_ATCC_1997 -------------
MLBr_01740|M.leprae_Br4923 SQPRSGEGGKSGN