For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VITAPSPLHRLSEQDLEKLAKEFDAIHDEVFAELGERDRHYIKTVISVQRQIIVAGRVLLLASRSRTAWV LGTACLSMAKILENMELGHNILHGQWDWMNDPDVHSSVWDWDTASSAKAWKHSHNYIHHTYTNILGKDKD LGYEIMRIDPHQKWRPGNLGQPLYNLLLMLLFEWGVAVHDTDIEGLIRGQKTFADLKDDFKEIGVKARQQ IVKDYLGWPMISAGAFALAQLALRGRLPRPPHSRLGNRLRRITGNRRVGTVAGLLDRHVAGVESTYLRTL AANALANVIRNVWAHAIIFCGHFPDQTWTFTEEEVENETRGGWYVRQLLGAANIEGSPLFHIISGNLGYQ VEHHLFPDMPSSRYAEIAPKVKDICERYELPYNSGPFGHQWFTVQRTIFRLAFPGGTPRPKPGPYRSPDR EHAPKGPSEAARFRGRVPAEHPDAGPEHASGGVAVQLPPRGND*
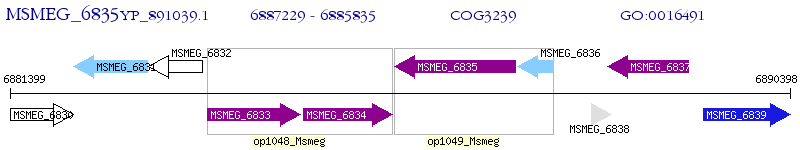
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6835 | - | - | 100% (464) | Fatty acid desaturase |
| M. smegmatis MC2 155 | MSMEG_1743 | - | e-106 | 45.72% (409) | Fatty acid desaturase |
| M. smegmatis MC2 155 | MSMEG_1886 | - | e-100 | 41.73% (405) | Fatty acid desaturase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3258c | - | 9e-98 | 39.72% (433) | linoleoyl-CoA desaturase |
| M. gilvum PYR-GCK | Mflv_5246 | - | 0.0 | 85.78% (464) | fatty acid desaturase |
| M. tuberculosis H37Rv | Rv3229c | - | 9e-98 | 39.72% (433) | linoleoyl-CoA desaturase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3548c | - | 1e-102 | 42.72% (405) | putative fatty acid desaturase |
| M. marinum M | MMAR_0336 | desA3_2 | 1e-103 | 45.16% (403) | linoleoyl-CoA desaturase, DesA3_2 |
| M. avium 104 | MAV_4192 | - | 1e-102 | 41.40% (430) | Fatty acid desaturase |
| M. thermoresistible (build 8) | TH_0650 | desA3 | 1e-100 | 42.72% (405) | POSSIBLE LINOLEOYL-CoA DESATURASE (DELTA(6)-DESATURASE) |
| M. ulcerans Agy99 | MUL_4785 | desA3_2 | 1e-101 | 44.67% (403) | linoleoyl-CoA desaturase, DesA3_2 |
| M. vanbaalenii PYR-1 | Mvan_1768 | - | 1e-104 | 42.53% (442) | fatty acid desaturase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3258c|M.bovis_AF2122/97 --------MAITDVDVFAH---LTDADIENLAAELDAIRRDVEESRGERD
Rv3229c|M.tuberculosis_H37Rv --------MAITDVDVFAH---LTDADIENLAAELDAIRRDVEESRGERD
MAV_4192|M.avium_104 --------MAITDVDVFAH---LTDADIENLAVELDAIRQDVEDSRGERD
TH_0650|M.thermoresistible__bu --------MAITDVPAYAH---LTEVDIDALAVELDAIRRDIEDSLGERD
Mvan_1768|M.vanbaalenii_PYR-1 --------MAITDVPQFAH---LTEADVENLGRELDAIRQDIEDSRGERD
MAB_3548c|M.abscessus_ATCC_199 --------MAISDIQEFAH---LTEADVEALGHELDAIRRDVEESLGAAD
MSMEG_6835|M.smegmatis_MC2_155 --------MVITAPSPLHR---LSEQDLEKLAKEFDAIHDEVFAELGERD
Mflv_5246|M.gilvum_PYR-GCK --------MTITAGSPLTR---LSEQELEKLAKEFDAIHDEVFAELGDRD
MMAR_0336|M.marinum_M MGNNNIATIAETADTATRANFALTPEQAEAFGRELDAIKERVIADLGEAD
MUL_4785|M.ulcerans_Agy99 MGNNNIATIAETADTATRANFALTPEQAEAFGRELDAIKERVIADLGEAD
:. : *: : : :. *:***: : . * *
Mb3258c|M.bovis_AF2122/97 ARYIRRTIAAQRALEVSGRLLLAGSSRRLAWWTGALTLGVAKIIENMEIG
Rv3229c|M.tuberculosis_H37Rv ARYIRRTIAAQRALEVSGRLLLAGSSRRLAWWTGALTLGVAKIIENMEIG
MAV_4192|M.avium_104 ARYIRRTIAAQRALEVTGRLLLAASSRRSAWWAGTVTLGVAKIIENMEIG
TH_0650|M.thermoresistible__bu ARYIRRTIATQRGLEVAGRLLLAASSRRSAWWAGAVTLAAAKIIENMEIG
Mvan_1768|M.vanbaalenii_PYR-1 ARYIRRTIAIQRGLEVTGRLLLAGSAKRRFWWAGTATLGVAKIIENMEIG
MAB_3548c|M.abscessus_ATCC_199 ARYIRRTIAAQRTLELTGRILLAASKKRSAWWAGAVTLGVAKIVENMEIG
MSMEG_6835|M.smegmatis_MC2_155 RHYIKTVISVQRQIIVAGRVLLLASRSRTAWVLGTACLSMAKILENMELG
Mflv_5246|M.gilvum_PYR-GCK RQYIKTVISAQRQIVVAGRVLLLASRSRTALVLGTACLGLAKILENMELG
MMAR_0336|M.marinum_M ADYIRRVIKAQRALEVGGRALLFAGILPPAWLAGTAMLGLSKILDNMEIG
MUL_4785|M.ulcerans_Agy99 ADYIRRVIKAQRALEVGGRALLFAGILPPTWLAGTAMLGLSKILDNMEIG
**: .* ** : : ** ** .. *: *. :**::***:*
Mb3258c|M.bovis_AF2122/97 HNVMHGQWDWMNDPEIHSSTWEWDMSGSSKHWRYTHNFVHHKYTNILGMD
Rv3229c|M.tuberculosis_H37Rv HNVMHGQWDWMNDPEIHSSTWEWDMSGSSKHWRYTHNFVHHKYTNILGMD
MAV_4192|M.avium_104 HNVMHGQWDWMNDPEIHSSTWEWDMSGSSKHWRYTHNFVHHKYTNILGMD
TH_0650|M.thermoresistible__bu HNVMHGQWDWMNDPEIHSSTWEWDMTGASKHWRFTHNFMHHKYTNILGMD
Mvan_1768|M.vanbaalenii_PYR-1 HNVMHGQWDWMNDPEIHSSSWEWDMSGASKHWRFTHNFMHHKYTNILGMD
MAB_3548c|M.abscessus_ATCC_199 HNVMHGQWDWMNDPEIHSSSWEWDMVDPAEHWKQTHNYIHHKYTNVLGMD
MSMEG_6835|M.smegmatis_MC2_155 HNILHGQWDWMNDPDVHSSVWDWDTASSAKAWKHSHNYIHHTYTNILGKD
Mflv_5246|M.gilvum_PYR-GCK HNILHGQWDWMNDPDIHSSVWDWDTASSAKAWKHSHNYIHHTYTNILGKD
MMAR_0336|M.marinum_M HNVMHGQYDWMRDPAISGRSFEWDTACPADQWRHSHNYMHHTHTNIVGMD
MUL_4785|M.ulcerans_Agy99 HNVMHDQYDWMRDPAISGRSFEWDTACPADQWRHSHNYMHHTHTNILGMD
**::*.*:***.** : . ::** .:. *: :**::**.:**::* *
Mb3258c|M.bovis_AF2122/97 DDVGYGMLRVTRDQRWKRYNIFNVVWNTILAIGFEWGVALQHLEIGKIFK
Rv3229c|M.tuberculosis_H37Rv DDVGYGMLRVTRDQRWKRYNIFNVVWNTILAIGFEWGVALQHLEIGKIFK
MAV_4192|M.avium_104 DDVGYGMLRVTRDQRWKRFNLFNLVWNTLLALLFEWGVGLQHVEIGKILK
TH_0650|M.thermoresistible__bu DDVGYGLLRVTRDQKWKPFNLGNLLYNTLLALAFEWGVGLQHVELGKIFK
Mvan_1768|M.vanbaalenii_PYR-1 DDVGYGVIRVTRDQRWRPMNLLNLFFNTLLSVAFEWGVGLQHLELGKIFK
MAB_3548c|M.abscessus_ATCC_199 DDVGYGLLRVTRDKKWKPFNLGNLLYNTLLMLLFEWGVGTQHLELGKVAQ
MSMEG_6835|M.smegmatis_MC2_155 KDLGYEIMRIDPHQKWRPGNLGQPLYNLLLMLLFEWGVAVHDTDIEGLIR
Mflv_5246|M.gilvum_PYR-GCK KDLGYEIMRIDPNQKWRPSNLGQPFYNFLLTVLFEWGVAVHDTDVESLFR
MMAR_0336|M.marinum_M RDIGYGIIRMSEDQPWQPYYLGNPVYAFLLMVLFQYGVALHELETERIRS
MUL_4785|M.ulcerans_Agy99 RDIGYGIIRMSEDQPWQPYYLGNPVYAFLLMVLFQYGVALHELETERIRS
*:** ::*: .: *: : : .: :* : *::**. :. : :
Mb3258c|M.bovis_AF2122/97 GRADREAAKTRLREFSAKAGRQVFKDYVAFPALTS---LSPGATYRSTLT
Rv3229c|M.tuberculosis_H37Rv GRADREAAKTRLREFSAKAGRQVFKDYVAFPALTS---LSPGATYRSTLT
MAV_4192|M.avium_104 RRMDHEEARQRIDEFLAKAGRQVVKDYVAFPALTS---LSPGATFLSTLK
TH_0650|M.thermoresistible__bu RRHDPDESRVRVREFLRKAGRQVFKDYVAYPALSS---LSPAASYRSTLT
Mvan_1768|M.vanbaalenii_PYR-1 GRDDRKATMGRVKEFGVKAGQQVVKDYVAYPAITS---LSPRATFRSTLT
MAB_3548c|M.abscessus_ATCC_199 GRADKEEFRVKRQAALTKMGKQVAKDYVAFPALSA---LSPAGSYRKTVT
MSMEG_6835|M.smegmatis_MC2_155 GQKTFADLKDDFKEIGVKARQQIVKDYLGWPMISAGAFALAQLALRGRLP
Mflv_5246|M.gilvum_PYR-GCK GEKKLKDLKEDFKSIGGKARQQIVKDYIGWPLVSAGAFGLAQLALRGRLD
MMAR_0336|M.marinum_M GEITLRDKRETLMEIWKKTRRQTLKDYVAFPLLAG---PFAPFVFAGNLT
MUL_4785|M.ulcerans_Agy99 GEITLRDKRETLTEIWKKTRRQTLKDYVAFPLLAG---PFAPFVFAGNLT
. * :* ***:.:* ::. . :
Mb3258c|M.bovis_AF2122/97 -------------------------------------------ANVVANV
Rv3229c|M.tuberculosis_H37Rv -------------------------------------------ANVVANV
MAV_4192|M.avium_104 -------------------------------------------ANAMANV
TH_0650|M.thermoresistible__bu -------------------------------------------ANAMANV
Mvan_1768|M.vanbaalenii_PYR-1 -------------------------------------------ANAVANV
MAB_3548c|M.abscessus_ATCC_199 -------------------------------------------ANLLANV
MSMEG_6835|M.smegmatis_MC2_155 RPPHSRLGNRLRRITGNRRVGTVAGLLDRHVAGVESTYLRTLAANALANV
Mflv_5246|M.gilvum_PYR-GCK QPSHSRIGKLLRRTTGNRRVGGVATLLDRYASGVESTYLRTLGADALANL
MMAR_0336|M.marinum_M -----------------------------------------------ANL
MUL_4785|M.ulcerans_Agy99 -----------------------------------------------ANL
**:
Mb3258c|M.bovis_AF2122/97 IRNVWSNAVIFCGHFPDGAEKFTKTDMIGEPKGQWYLRQMLGSANFNAGP
Rv3229c|M.tuberculosis_H37Rv IRNVWSNAVIFCGHFPDGAEKFTKTDMIGEPKGQWYLRQMLGSANFNAGP
MAV_4192|M.avium_104 IRNVWANAVIFCGHFPDGAEKFTKTDMIGETKGQWYLRQMLGSANFEAGP
TH_0650|M.thermoresistible__bu IRNVWSNAVIFCGHFPDGAEKFTKTDMVGESKGQWYLRQMLGSANFEGGW
Mvan_1768|M.vanbaalenii_PYR-1 IRNVWANAVIFCGHFPDGAEKFTKTDMVGESRGQWYLRQMLGSANFEGGP
MAB_3548c|M.abscessus_ATCC_199 IRNIWSNAVIFCGHFPDGAEKFTKQDLENETQGQWYLRQMLGSANFDAGP
MSMEG_6835|M.smegmatis_MC2_155 IRNVWAHAIIFCGHFPDQTWTFTEEEVENETRGGWYVRQLLGAANIEGSP
Mflv_5246|M.gilvum_PYR-GCK IRNVWAHAIIFCGHFPDQTWTFTEEEVENETRGGWYVRQLLGAANIEGSP
MMAR_0336|M.marinum_M MRNVWSYMIIFCGHFPDGTQEFSVEETQNESRGQWYFRQVLGSANLTGGK
MUL_4785|M.ulcerans_Agy99 MRNVWSYMIIFCGHFPDGTQEFSVEETQNESRGQWYFRQVLGSANLTGGK
:**:*: :******** : *: : .*.:* **.**:**:**: ..
Mb3258c|M.bovis_AF2122/97 ALRFMSGNLCHQIEHHLYPDLPSNRLHEISVRVREVCDRYDLPYTTGSFL
Rv3229c|M.tuberculosis_H37Rv ALRFMSGNLCHQIEHHLYPDLPSNRLHEISVRVREVCDRYDLPYTTGSFL
MAV_4192|M.avium_104 VLRFMSGNLCHQIEHHLYPDLPSNRLHEISVRVRQVCDKYDLPYTTGSFL
TH_0650|M.thermoresistible__bu LLRFMSGNLCHQIEHHLYPDLPSNRLHEASIRVREVCDKYDLPYTTGPFL
Mvan_1768|M.vanbaalenii_PYR-1 LLRFMSGNLSQQIEHHLFPDLPSNRYYEISLRVRELCDKYDLPYTTGNFL
MAB_3548c|M.abscessus_ATCC_199 AMAFMSGNLCHQIEHHLFPDLPSNRLSAVSVRVRELCDKYDLPYTSGPFL
MSMEG_6835|M.smegmatis_MC2_155 LFHIISGNLGYQVEHHLFPDMPSSRYAEIAPKVKDICERYELPYNSGPFG
Mflv_5246|M.gilvum_PYR-GCK LFHVLSGNLGYQVEHHLFPDMPSSRYAEIAPKVKDICERYELPYNSGRFG
MMAR_0336|M.marinum_M TFHLLSGNLSHQIEHHLFPDIPARRYAEIAPEVQEICERYGIPYNRGPLL
MUL_4785|M.ulcerans_Agy99 TFHLLSGNLSHQREHHLFPDIPARRYAEIAPEVQEICELYGIPYNRGPLL
: .:**** * ****:**:*: * : .*:::*: * :**. * :
Mb3258c|M.bovis_AF2122/97 VQYGKTWRTLAKLSLPDKYLRDNADDAPETRSERMFAGLGPGFAGADPVT
Rv3229c|M.tuberculosis_H37Rv VQYGKTWRTLAKLSLPDKYLRDNADDAPETRSERMFAGLGPGFAGADPVT
MAV_4192|M.avium_104 FQYAKTWRTLAKLSLPNKYLRDDADNAPETRSERMFAELDPDFLGTDPAT
TH_0650|M.thermoresistible__bu VQYAKTWRTLAKLSLPDRFLKATADNAPETRSERMFAELGPGFAGVDPAT
Mvan_1768|M.vanbaalenii_PYR-1 VQYGKSWRTIAKLSLPDRFLTDTADDAPETRSERMFDGLEP---------
MAB_3548c|M.abscessus_ATCC_199 VQYAKAWRTIAKLSLPDKYLTYTADNAQETRSVKMFDELEP---------
MSMEG_6835|M.smegmatis_MC2_155 HQWFTVQRTIFRLAFPGGTPRPKPG--PYRSPDREHAPKGP---------
Mflv_5246|M.gilvum_PYR-GCK KQWFTVHRTIFRLALPGGTPRPKPG--PYRSPERGRSPQGV---------
MMAR_0336|M.marinum_M RQFGTVVRKIVKLAFRG--------------SDRQ---------------
MUL_4785|M.ulcerans_Agy99 RQFGTVVRQIVKLAFRG--------------SDRQ---------------
*: . * : :*:: . . :
Mb3258c|M.bovis_AF2122/97 GRRRGLKTAIAAVRGRRRSKRMAKSVT------EPDDLAA----------
Rv3229c|M.tuberculosis_H37Rv GRRRGLKTAIAAVRGRRRSKRMAKSVT------EPDDLAA----------
MAV_4192|M.avium_104 GRRRGLKSAIAAVRGWRRGKRASVARR------TGGELAA----------
TH_0650|M.thermoresistible__bu GRRRGLKSAFAAVRNWRRDKQAAKAXXXGELYPRPGTLRRDVGVRLRRGG
Mvan_1768|M.vanbaalenii_PYR-1 SERRGLRSAIAAVRKRRRAKAAEKAAV---------KLAA----------
MAB_3548c|M.abscessus_ATCC_199 HERRGLKSAFAAVRARRKARHKV---------------------------
MSMEG_6835|M.smegmatis_MC2_155 SEAARFRGRVPAEHPDAGPEHASGGVAVQLPPRGND--------------
Mflv_5246|M.gilvum_PYR-GCK SEAERFRSRVPAEHPAAGPEHESGGVAVAPPARGND--------------
MMAR_0336|M.marinum_M -EAHALA-------------------------------------------
MUL_4785|M.ulcerans_Agy99 -EAHALA-------------------------------------------
. :
Mb3258c|M.bovis_AF2122/97 ---------------------------------------------
Rv3229c|M.tuberculosis_H37Rv ---------------------------------------------
MAV_4192|M.avium_104 ---------------------------------------------
TH_0650|M.thermoresistible__bu VPGPAGLWSGEHDRRGHRPPRSGPQRIRRPRGHAVLRRRDRAVRR
Mvan_1768|M.vanbaalenii_PYR-1 ---------------------------------------------
MAB_3548c|M.abscessus_ATCC_199 ---------------------------------------------
MSMEG_6835|M.smegmatis_MC2_155 ---------------------------------------------
Mflv_5246|M.gilvum_PYR-GCK ---------------------------------------------
MMAR_0336|M.marinum_M ---------------------------------------------
MUL_4785|M.ulcerans_Agy99 ---------------------------------------------