For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SHGDGAAADPVTDPAVVESLRDRLWDAVIEADEDAAVDVVFGALEDGVPGEDLLLDVIAAVQRRVGVEWA DNRLTVAAEHAATAINDRVIAALVRHPAVARQTTTGRVAVACADGEWHALPARLLAEVLRLRGWRVDFLG AQVPTPHLIAHLHQHGPDAVALSCSISTRLPTAHAAITACQGAGVPVLVGGAGFGPDGRYARLLGADAWA PDARAAAAQLADGLALTRRAPRDHLFDELPHLADQEYTMVSTTAGRLVRDTVSGLEKRVPAMASYSQLQR QHTSTDVAHIVDFLATALYVDDPELFTGFIAWTAEVLDARGVPVDSLQPTLELLGAQLQDFPRTQRMLSA ARDMLHATYARRNG*
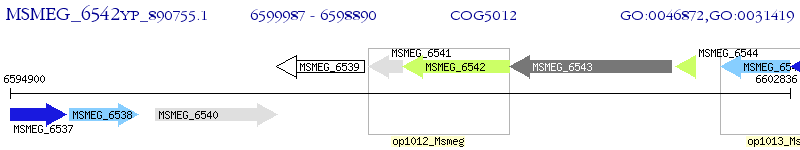
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6542 | - | - | 100% (365) | B12-binding domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1030 | - | 1e-129 | 67.75% (338) | cobalamin B12-binding domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4825 | metH | 6e-05 | 28.57% (91) | 5-methyltetrahydrofolate--homocysteine methyltransferase, |
| M. avium 104 | MAV_2379 | metH | 2e-06 | 24.02% (204) | B12-dependent methionine synthase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0395 | metH | 4e-05 | 28.57% (91) | B12-dependent methionine synthase |
| M. vanbaalenii PYR-1 | Mvan_5806 | - | 1e-134 | 71.00% (331) | cobalamin B12-binding domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4825|M.marinum_M ---------------------------------MSAFEPNIRPDCTDALT
MUL_0395|M.ulcerans_Agy99 MWVERRCNGSPVPRRYPPRSAESRTPSATEGINVSAFEPNIRPDCTDALT
MAV_2379|M.avium_104 ------------------MTAPVTTP------ASDSFAPNIRPDCTDELT
Mflv_1030|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5806|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6542|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4825|M.marinum_M AALGQRIMVIDGAMGTAIQRDRPDEAGYRGERFKDWPSDLVGNNDLLSLT
MUL_0395|M.ulcerans_Agy99 ATLGQRIMVIDGAMGTAIQRDRPDEAGYRGERFKDWPSDLVGNNDLLSLT
MAV_2379|M.avium_104 AALRQRIMVIDGAMGTAIQRDRPDEAGYRGERFKDWPSDLVGNNDLLNLT
Mflv_1030|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5806|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6542|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4825|M.marinum_M QPQIIEGIHREYLQAGADILETNTFNANAVSLSDYDMAELAYELNYAGAA
MUL_0395|M.ulcerans_Agy99 QPQIIEGIHREYLQAGADILETNTFNANAVSLSDYDMAELAYELNYAGAA
MAV_2379|M.avium_104 QPQIIEGIHREYLEAGADILETNTFNANAISLSDYGMEELSYELNYAGAA
Mflv_1030|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5806|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6542|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4825|M.marinum_M LARAAADEFSTPEKPRYVAGALGPTTRTASISPDVNDPGARNVSYDQLVA
MUL_0395|M.ulcerans_Agy99 LARAAADEFSTPEKPRYVAGALGPTTRTASISPDVNDPGARNVSYDQLVA
MAV_2379|M.avium_104 LARKAADEFSTPDKPRYVAGALGPTSRTASISPDVNDPGARNVTYDELVA
Mflv_1030|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5806|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6542|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4825|M.marinum_M AYLEAANGLVDGGADIIIIETIFDSLNAKAAVFAVETLFEDRGRRWPVII
MUL_0395|M.ulcerans_Agy99 AYLEAANGLVDGGADIIIIETIFDSLNAKAAVFAVETLFEDRGRRWPVII
MAV_2379|M.avium_104 AYLEAANGLVDGGVDLLIVETIFDTLNAKAAVFALETLFEQRGRRWPVII
Mflv_1030|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5806|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6542|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4825|M.marinum_M SGTITDASGRTLSGQVTEAFWNAIRHAKPIAVGLNCALGAPEMRPYIAEM
MUL_0395|M.ulcerans_Agy99 SGTITDASGRTLSGQVTEAFWNAIRHAKPIAVGLNCALGAPEMRPYIAEM
MAV_2379|M.avium_104 SGTITDASGRTLSGQVTEAFWNSIRHAKPIAVGLNCALGAPEMRPYIAEV
Mflv_1030|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5806|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6542|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4825|M.marinum_M ARIADTFVSCYPNAGLPNAFGEYDETPEHQAGYIAEFADAGLVNLVGGCC
MUL_0395|M.ulcerans_Agy99 ARIADTFVSCYPNAGLPNAFGEYDETPEHQAGYIAEFADAGLVNLVGGCC
MAV_2379|M.avium_104 SRIADTFVSCYPNAGLPNAFGEYDESPERQASYIADFAEAGLVNIVGGCC
Mflv_1030|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5806|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6542|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4825|M.marinum_M GTAPPHIAEIAKVVEGKAPRELPEIPVATRLSGLEPLNISDDSLFVNIGE
MUL_0395|M.ulcerans_Agy99 GTAPPHIAEIAKVVEGKAPRELPEIPVATRLSGLEPLNITDDSLFENIGE
MAV_2379|M.avium_104 GTAPPHIAEIAKVVEGKPPREVPQIPVATRLSGLEPLNITDDSLFVNIGE
Mflv_1030|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5806|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6542|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4825|M.marinum_M RTNITGSARFRNLIKAEDYDTALSVALQQVEVGAQVIDINMDEGMIDGVA
MUL_0395|M.ulcerans_Agy99 RTNITGSARFRNLIKAEDYDTALSVALQQVEVGAQVIDINMDEGMIDGVA
MAV_2379|M.avium_104 RTNITGSARFRNLIKAEDYDTALSVALQQVEVGAQVIDINMDEGMIDGVA
Mflv_1030|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5806|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6542|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4825|M.marinum_M AMDRFTKLIAAEPDISRVPVMIDSSKWEVIEAGLKNVQGKPIVNSISMKE
MUL_0395|M.ulcerans_Agy99 AMDRFTKLIAAEPDISRVPVMIDSSKWEVIEAGLKNVQGKPIVNSISMKE
MAV_2379|M.avium_104 AMDRFTRLIAAEPDISRVPVMIDSSKWEVIEAGLKNVQGKPIVNSISLKE
Mflv_1030|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5806|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6542|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4825|M.marinum_M GEEKFVREARLCRKYGAAVVVMAFDEKGQADNLERRKEICGRAYRILTEE
MUL_0395|M.ulcerans_Agy99 GEEKFVREARLCRKYGAAVVVMAFDEKGQADNLERRKEICGRAYRILTEE
MAV_2379|M.avium_104 GEEKFVREARLCRKYGAAVVVMAFDEQGQADNLERRKQICARAYRVLTEE
Mflv_1030|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5806|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6542|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4825|M.marinum_M VGFPAEDIIFDPNCFALATGIEEHATYGIDFIEACAWIKENLPGVHISGG
MUL_0395|M.ulcerans_Agy99 VGFPAEDIIFDPNCFALATGIEEHATYGIDFIEACAWIKENLPGVHISGG
MAV_2379|M.avium_104 VGFPPEDIIFDPNCFALATGIEEHATYGIDFIEACAWIKENLPGVHLSGG
Mflv_1030|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5806|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6542|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4825|M.marinum_M ISNVSFSFRGNNPVREAIHAVFLFHAIKAGLDMGIVNAGALVPYDSIDPE
MUL_0395|M.ulcerans_Agy99 ISNVSFSFRGNNPVREAIHAVFLFHAIKAGLDMGIVNAGALVPYDSIDPE
MAV_2379|M.avium_104 ISNVSFSFRGNNPVREAIHSVFLFHAIKAGLDMGIVNAGALVPYDSIDPE
Mflv_1030|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5806|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6542|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4825|M.marinum_M LRERIEDVVLNRREDAAERLLEIAERFN-TKGKSEDPAAAEWRSLPVRER
MUL_0395|M.ulcerans_Agy99 LRERIEDVVLNRREDAAERLLEIAERFN-TKGKPEDPAAAEWRSLPVRER
MAV_2379|M.avium_104 LRDRIEDVVLNRREDAAERLLEIAERFNKSADASEDSAAAEWRGLPVRER
Mflv_1030|M.gilvum_PYR-GCK -----------------------------MSALNPD-------CLALQER
Mvan_5806|M.vanbaalenii_PYR-1 --------------------------------MSVD-------ASRARER
MSMEG_6542|M.smegmatis_MC2_155 --------------------------MSHGDGAAADPVTDPAVVESLRDR
* ::*
MMAR_4825|M.marinum_M ITHALVKGIDAHVDADTEELRAEIAAAGGRPIEVIEGPLMDGMNVVGDLF
MUL_0395|M.ulcerans_Agy99 ITHALVKGIDAHVDADTEELRAEIAAAGGRLIEVIEGPLMDGMNVVGDLF
MAV_2379|M.avium_104 ITHALVKGIDAHVDEDTEELRAEIAAAGGRPIEVIEGPLMDGMNVVGDLF
Mflv_1030|M.gilvum_PYR-GCK LWAAVMD----GDEYTASTVVFGALDAGLDPEDVLLDVIAVVQRRVGTEW
Mvan_5806|M.vanbaalenii_PYR-1 LWAAVLE----ADEFAAATTVFGAVDGGMPPEDVLLDVIASVQRRVGSEW
MSMEG_6542|M.smegmatis_MC2_155 LWDAVIE----ADEDAAVDVVFGALEDGVPGEDLLLDVIAAVQRRVGVEW
: *::. : : * ::: . : . ** :
MMAR_4825|M.marinum_M GAGKMFLPQVVKSARVMKKAVAYLLPFIEAEKAQSGSTEQDKTNGIIIMA
MUL_0395|M.ulcerans_Agy99 GAGKMFLPQVVKSARVMKKAVAYLLPFIEAEKAQSGSTEQDKTNGIIIMA
MAV_2379|M.avium_104 GSGKMFLPQVVKSARVMKKAVAYLLPFIEAEKEESGVSGSKDTNGTIVMA
Mflv_1030|M.gilvum_PYR-GCK AANRITVAQEHAATAINDRVIAALAHHPKACRR--------PDAGRVTVA
Mvan_5806|M.vanbaalenii_PYR-1 AANRITVAQEHAATAINDRVIAALAHHPACSRA--------DGAGRVTVA
MSMEG_6542|M.smegmatis_MC2_155 ADNRLTVAAEHAATAINDRVIAALVRHPAVARQ--------TTTGRVAVA
. .:: :. :: : .:.:* * . : * : :*
MMAR_4825|M.marinum_M TVKGDVHDIGKNIVGVVLQCNNYTVVDLGVMVPAEKILAAAREYDADIIG
MUL_0395|M.ulcerans_Agy99 TVKGDVHDIGKNIVGVVLQCNNYTVVDLGVMVPAEKILAAAREYDADIIG
MAV_2379|M.avium_104 TVKGDVHDIGKNIVGVVLQCNNFEVIDLGVMVPAQKILDAAKEHDADIIG
Mflv_1030|M.gilvum_PYR-GCK CVDGEWHALPARLLAEVLRLRGWQVDFLGAQVPTPHLIAHVHQHGPVAVA
Mvan_5806|M.vanbaalenii_PYR-1 CVDGEWHALPARLLAEVLRLRGWHVDFLGAQVPTPHLIAHLHQHGPEAVA
MSMEG_6542|M.smegmatis_MC2_155 CADGEWHALPARLLAEVLRLRGWRVDFLGAQVPTPHLIAHLHQHGPDAVA
..*: * : .::. **: ..: * **. **: ::: :::.. :.
MMAR_4825|M.marinum_M LSGLITPSLDEMVNFAAAMEREGMQIPLLIGGATTSRAHTAVKVAPRRSG
MUL_0395|M.ulcerans_Agy99 LSGLITPSLDEMVNFAAAMEREGMQIPLLIGGATTSRAHTAVKVAPRRSG
MAV_2379|M.avium_104 LSGLITPSLDEMSNFAAEMEREGLQIPLLIGGATTSRAHTAVKISPRRSG
Mflv_1030|M.gilvum_PYR-GCK LSCSIPTRLPTAH--AAITACQAAGVPVLAGGAAFG----FDGRFARLLG
Mvan_5806|M.vanbaalenii_PYR-1 LSCMIPTRLPTAH--AAITACQAAGTAVLAGGAAFG----ADGRFARQLG
MSMEG_6542|M.smegmatis_MC2_155 LSCSISTRLPTAH--AAITACQGAGVPVLVGGAGFG----PDGRYARLLG
** *.. * ** :. .:* *** . .* *
MMAR_4825|M.marinum_M PVVWVKDASRSVPVAAALLDDKQRPGLLEATAADYAALRERHSQKNERPM
MUL_0395|M.ulcerans_Agy99 PVVWVKDASRSVPVAAALLDDKQRPGLLEATAADYAALRERHSQKNERPM
MAV_2379|M.avium_104 PVVWVKDASRSVPVAAALLDDKQRPALLEATEKDYASLRERHAQKNERPM
Mflv_1030|M.gilvum_PYR-GCK ADAWASDAR----TAADLLRGGVRPAQSTATHQPIDDL--PHLADQEYTM
Mvan_5806|M.vanbaalenii_PYR-1 ADAWAPDAR----AAAELLGRGLRPARTSPTHQPIDDL--PHLADQEYTM
MSMEG_6542|M.smegmatis_MC2_155 ADAWAPDAR----AAAAQLADGLALTRRAPRDHLFDEL--PHLADQEYTM
. .*. ** .** * . * * .:* .*
MMAR_4825|M.marinum_M LTLEKARANRTPIDWDGYTPPLPAQGLGVREFHDYDLAELREYIDWQPFF
MUL_0395|M.ulcerans_Agy99 LTLEKARANRTPIDWDGYTPPLPAQGLGVREFHDYDLAELREYIDWQPFF
MAV_2379|M.avium_104 VTLEKARANRTPIDWDGYTPPVPAIGAGIREFQDYDLAELREYIDWQPFF
Mflv_1030|M.gilvum_PYR-GCK VRSTATTLVQT---------------------------------------
Mvan_5806|M.vanbaalenii_PYR-1 VKSAGAQLIQT---------------------------------------
MSMEG_6542|M.smegmatis_MC2_155 VSTTAGRLVRD---------------------------------------
: :
MMAR_4825|M.marinum_M NAWEMKGRFPDILNNPATGEAARKLYNDAQEMLDTLIREKWLTANGVIGF
MUL_0395|M.ulcerans_Agy99 NAWEMKGRFPDILNNPATGEAARKLYNDAQEMLDTLIREKWLTANGVIGF
MAV_2379|M.avium_104 NAWEMKGRFPDILNNPATGEAARKLYDDAQEMLDTLIKEKWLTANAVIGF
Mflv_1030|M.gilvum_PYR-GCK TVEELEDRYPAMRD--YNPAQRERTAEDIAHIVAFLS-------------
Mvan_5806|M.vanbaalenii_PYR-1 TLWELEQRFPAMRG--YTEMQRQHTIEDLAHIVDFLT-------------
MSMEG_6542|M.smegmatis_MC2_155 TVSGLEKRVPAMAS--YSQLQRQHTSTDVAHIVDFLA-------------
. :: * * : . . .: * .:: *
MMAR_4825|M.marinum_M FPANAVG---DDIEVYTDETRAEVLTTLHNLRQQGEHRAGIPNRSLGDFI
MUL_0395|M.ulcerans_Agy99 FPANAVG---DDIEVYTDETRAEVLTTLHNLRQQGEHRAGIPNRSLGDFI
MAV_2379|M.avium_104 FPANAIGPGFEDIELYTDDTRTEVLTTLHNLRQQGEHRDGIPNRSLGDYV
Mflv_1030|M.gilvum_PYR-GCK ------------TALYTDDD--DLFTEFVCWTAEILVARAVPAESLYPAL
Mvan_5806|M.vanbaalenii_PYR-1 ------------TALYVDDD--DLFTGFITWTADILAARDVPAASLIPAL
MSMEG_6542|M.smegmatis_MC2_155 ------------TALYVDDP--ELFTGFIAWTAEVLDARGVPVDSLQPTL
:*.*: :::* : : :* ** :
MMAR_4825|M.marinum_M APKDTGLADYVGAFAVTAGLGSQTKIVEFKEALDDYSAILLESIADRLAE
MUL_0395|M.ulcerans_Agy99 APKDTGLADYVGAFAVTAGLGSQTKIVEFKEALDDYSAILLESIADRLAE
MAV_2379|M.avium_104 APKETGHRDYIGAFAVTAGLGSQEKIAEFKAALDDYSAILLESIADRLAE
Mflv_1030|M.gilvum_PYR-GCK DSLSGQLREFP------------------------RAQRLLTAAHAALHQ
Mvan_5806|M.vanbaalenii_PYR-1 ESLAGQLPEFP------------------------RTQRLLKAAHAAFPP
MSMEG_6542|M.smegmatis_MC2_155 ELLGAQLQDFP------------------------RTQRMLSAARDMLHA
:: : :* : :
MMAR_4825|M.marinum_M AFAERMHQRVREEFWGYQPDEQLDNEALIGERYVGIRPAPGYPACPEHTE
MUL_0395|M.ulcerans_Agy99 AFAERMHQRVREEFWGYQPDEQLDNEALIGERYVGIRPAPGYPACPEHTE
MAV_2379|M.avium_104 AFAERMHQRVRKEFWGYQPDEQLDNDALIDEKYRGIRPAPGYPACPEHTE
Mflv_1030|M.gilvum_PYR-GCK STTTNPPLFA----------------------------------------
Mvan_5806|M.vanbaalenii_PYR-1 TPTT-PSLPV----------------------------------------
MSMEG_6542|M.smegmatis_MC2_155 TYARRNG-------------------------------------------
: :
MMAR_4825|M.marinum_M KTTLFELMDVTKRTGIELTESMAMWPGAAVSGWYFSHPQSQYFVVGRMAQ
MUL_0395|M.ulcerans_Agy99 KTTLFELMDVTKRTGIELTESMAMWPGAAVSGWYFSHPQSQYFVVGRMAQ
MAV_2379|M.avium_104 KVTLWKLMDVKERTGIELTESMAMWPGAAVSGWYFSHPQSQYFVVGRLAQ
Mflv_1030|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5806|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6542|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4825|M.marinum_M DQVADYAKRKGWTLPEAERWLGPNLGYNPED
MUL_0395|M.ulcerans_Agy99 DQVADYAKRKGWTLPEAERWLGPNLGYNPED
MAV_2379|M.avium_104 DQVADYAKRKGWTLAEAERWLAPNLGYNPED
Mflv_1030|M.gilvum_PYR-GCK -------------------------------
Mvan_5806|M.vanbaalenii_PYR-1 -------------------------------
MSMEG_6542|M.smegmatis_MC2_155 -------------------------------