For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STPGTGAAGATSGVAVELVDLTRSYGSVKALDGLSLRMEPGELVALLGPSGCGKTTALRILAGLDEATSG TVLVDGRDVSSVPANKRDMGMVFQAYSLFPHLTVLDNVAFGLKMRGKSKRDRLSTAADMLDLVGLGAHKN KYANELSGGQQQRVALARALAIAPRVLLLDEPLSALDAKVRAQLRDEIRRVQLEVGTTTLFVTHDQEEAL AVADRVGVMSQGKLEQLAAPAELYANPATPFVAEFVGLHNKIPAEVTDGKARVLGATVPTLAGSVGSGAG TAMVRPESVTVQADPDGNATVASVAFLGPISRVYVNLPDGPVIHAQLPSSQARAFEAGQTVSVGLEPAPV LVV*
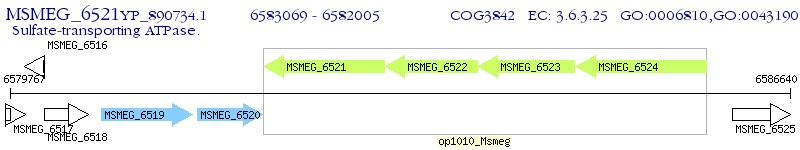
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6521 | - | - | 100% (354) | sulfate/thiosulfate import ATP-binding protein CysA 2 |
| M. smegmatis MC2 155 | MSMEG_6671 | - | 1e-83 | 48.09% (341) | sulfate/thiosulfate import ATP-binding protein CysA |
| M. smegmatis MC2 155 | MSMEG_0662 | - | 2e-71 | 40.88% (362) | putrescine transport ATP-binding protein PotG |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2419c | cysA1 | 3e-66 | 43.07% (339) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. gilvum PYR-GCK | Mflv_1083 | - | 1e-141 | 77.06% (340) | ABC transporter related |
| M. tuberculosis H37Rv | Rv2397c | cysA1 | 3e-66 | 43.07% (339) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. leprae Br4923 | MLBr_01424 | - | 7e-57 | 45.28% (265) | putative binding-protein-dependent transport protein |
| M. abscessus ATCC 19977 | MAB_1655 | - | 1e-65 | 46.01% (313) | sulfate ABC transporter, ATP-binding protein SysA |
| M. marinum M | MMAR_3715 | cysA1 | 2e-66 | 43.02% (351) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. avium 104 | MAV_1785 | - | 3e-65 | 43.06% (346) | sulfate/thiosulfate import ATP-binding protein CysA |
| M. thermoresistible (build 8) | TH_3667 | - | 1e-67 | 44.29% (359) | PUTATIVE ferric cations import ATP-binding protein FbpC |
| M. ulcerans Agy99 | MUL_3658 | cysA1 | 1e-65 | 42.45% (351) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. vanbaalenii PYR-1 | Mvan_5739 | - | 1e-145 | 78.07% (342) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1083|M.gilvum_PYR-GCK ------------------MTN-------------RGVGVQFDELTRVYG-
Mvan_5739|M.vanbaalenii_PYR-1 ----------------MSVGT-------------NGVGVQLDELTRVYG-
MSMEG_6521|M.smegmatis_MC2_155 ---------MSTPGTGAAGAT-------------SGVAVELVDLTRSYG-
TH_3667|M.thermoresistible__bu VIKTMKTKPADTSGAGPAVSNPAVSNPDVSAPQLRGADVEVQGLHKSFG-
Mb2419c|M.bovis_AF2122/97 ------------------MT----------------YAIVVADATKRYG-
Rv2397c|M.tuberculosis_H37Rv ------------------MT----------------YAIVVADATKRYG-
MAV_1785|M.avium_104 ------------------MTDAGTD--------RGDIAITVRDAYKRYG-
MMAR_3715|M.marinum_M ------------MKKETSLTGPG----------TGGHAIVVRDAFKQYG-
MUL_3658|M.ulcerans_Agy99 ------------------MTGPG----------TGGHAIVVRDAFKQYG-
MAB_1655|M.abscessus_ATCC_1997 ------------------MTAEN--------------TITVRGANKHYG-
MLBr_01424|M.leprae_Br4923 -----------------------------------MASVTFEQATRQFPD
: . : :
Mflv_1083|M.gilvum_PYR-GCK -ATRALDGLTLHIEPGELVALLGPSGGGKTTALRILAGLDEATSGTVSVG
Mvan_5739|M.vanbaalenii_PYR-1 -TTRALDGFTLHIEPGELVALLGPSGCGKTTALRILAGLDEATSGTVAVG
MSMEG_6521|M.smegmatis_MC2_155 -SVKALDGLSLRMEPGELVALLGPSGCGKTTALRILAGLDEATSGTVLVD
TH_3667|M.thermoresistible__bu -AKAVLRGVDLTVSAGTLTAVLGPSGCGKTTLLRILAGFDDPDAGTVRIG
Mb2419c|M.bovis_AF2122/97 -DFVALDHVDFVVPTGSLTALLGPSGSGKSTLLRTIAGLDQPDTGTITIN
Rv2397c|M.tuberculosis_H37Rv -DFVALDHVDFVVPTGSLTALLGPSGSGKSTLLRTIAGLDQPDTGTITIN
MAV_1785|M.avium_104 -DFVALDHVDFVVPTGSLTALLGPSGSGKSTLLRTIAGLDQPDTGTVTIY
MMAR_3715|M.marinum_M -DFVALDHVDFVVPAGSLTALLGPSGSGKSTLLRTIAGLDQPDSGNITIN
MUL_3658|M.ulcerans_Agy99 -DFVALDHVDFVVPAGSLTALLGPSGSGKSTLLRTIAGLDQPDSGNITIN
MAB_1655|M.abscessus_ATCC_1997 -DFAALDNIDFDVPSGSLTALLGPSGSGKSTLLRSIAGLDNPDSGTITIN
MLBr_01424|M.leprae_Br4923 TDRPALDRLDLDVDDGEFVVVVGPSGCGKTTSLRMVAGLETLDSGCIRIG
.* . : : * :..::**** **:* ** :**:: :* : :
Mflv_1083|M.gilvum_PYR-GCK GVDVG----RVPANKRDMGMVFQAYSLFPHLTVLDNVAFGLKMRGKGKSD
Mvan_5739|M.vanbaalenii_PYR-1 GVDVS----KVPANKRDMGMVFQAYSLFPHLTVLDNVAFGLKMRGKAKRD
MSMEG_6521|M.smegmatis_MC2_155 GRDVS----SVPANKRDMGMVFQAYSLFPHLTVLDNVAFGLKMRGKSKRD
TH_3667|M.thermoresistible__bu GHTLAGGGAPVPAHRRRVGLMPQEGALFPHLSVAENIAFGLRRTRRAG--
Mb2419c|M.bovis_AF2122/97 GRDVT----RVPPQRRGIGFVFQHYAAFKHLTVRDNVAFGLKIRKRPKAE
Rv2397c|M.tuberculosis_H37Rv GRDVT----RVPPQRRGIGFVFQHYAAFKHLTVRDNVAFGLKIRKRPKAE
MAV_1785|M.avium_104 GRDVT----RVPPQRRGIGFVFQHYAAFKHLTVRDNVAYGLKVRKRPKAE
MMAR_3715|M.marinum_M GRDVT----RVPPQRRGIGFVFQHYAAFKHLTVRDNVAYGLKIRKRPKAE
MUL_3658|M.ulcerans_Agy99 GRDVT----RVPPQRRGIGFVFQHYAAFKHLTVRDNVAYGLKIRKRPKAE
MAB_1655|M.abscessus_ATCC_1997 GRDVT----GVPPQRRDIGFVFQHYAAFKHLTVWENVAFGLKIRKKPKAE
MLBr_01424|M.leprae_Br4923 DRDVT----HVDPKDRDVAMVFQNYALYPHMTVAQNLGFALKISKISKAE
. : * .: * :.:: * : : *::* :*:.:.*:
Mflv_1083|M.gilvum_PYR-GCK RLSRAADMLDLVGLGELGGRYANELSGGQQQRVALARALAIRPRVLLLDE
Mvan_5739|M.vanbaalenii_PYR-1 RLSRAADMLELVGLGELGGRYAKELSGGQQQRVALARALAIRPRVLLLDE
MSMEG_6521|M.smegmatis_MC2_155 RLSTAADMLDLVGLGAHKNKYANELSGGQQQRVALARALAIAPRVLLLDE
TH_3667|M.thermoresistible__bu RADTVRYWLEVVGLEGAAEARPHELSGGQQQRVALARALAAEPRVLLLDE
Mb2419c|M.bovis_AF2122/97 IKAKVDNLLQVVGLSGFQSRYPNQLSGGQRQRMALARALAVDPEVLLLDE
Rv2397c|M.tuberculosis_H37Rv IKAKVDNLLQVVGLSGFQSRYPNQLSGGQRQRMALARALAVDPEVLLLDE
MAV_1785|M.avium_104 IKAKVDNLLEVVGLSGFQGRYPNQLSGGQRQRMALARALAVDPQVLLLDE
MMAR_3715|M.marinum_M IRDKVDNLLEVVGLSGFHGRYPNQLSGGQRQRMALARALAVDPEVLLLDE
MUL_3658|M.ulcerans_Agy99 IRDKVDNLLEVVGLSGFHGRYPDQLSGGQRQRMALARALAVDPEVLLLDE
MAB_1655|M.abscessus_ATCC_1997 IKKKVDDLLEVVGLSGFHTRYPAQLSGGQRQRMALARALAVDPQVLLLDE
MLBr_01424|M.leprae_Br4923 IHDRILVVAKLLDLQPYLDRKPKDLSGGQRQRVAMGRAIVRRPQVFLMDE
.::.* . :*****:**:*:.**:. *.*:*:**
Mflv_1083|M.gilvum_PYR-GCK PLSALDAKVRTQLRDEIRRVQLEVGTTTLFVTHDQEEALAVADRVGVMNQ
Mvan_5739|M.vanbaalenii_PYR-1 PLSALDAKVRTQLRDEIRRVQLEVGTTTLFVTHDQEEALAVADRVGVMNE
MSMEG_6521|M.smegmatis_MC2_155 PLSALDAKVRAQLRDEIRRVQLEVGTTTLFVTHDQEEALAVADRVGVMSQ
TH_3667|M.thermoresistible__bu PFVALDAGLRVRVREEIAAILRAAGTTAVLVTHDQSEALSLADSVALLMD
Mb2419c|M.bovis_AF2122/97 PFGALDAKVREELRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAVLHK
Rv2397c|M.tuberculosis_H37Rv PFGALDAKVREELRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAVLHK
MAV_1785|M.avium_104 PFGALDAKVREDLRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAVLNQ
MMAR_3715|M.marinum_M PFGALDAKVREDLRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAVLNS
MUL_3658|M.ulcerans_Agy99 PFGALDAKVREDLRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAVLNS
MAB_1655|M.abscessus_ATCC_1997 PFGALDAKVREDLRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAVLNK
MLBr_01424|M.leprae_Br4923 PLSNLDAKLRAQTRNQIAELQCQLGTTTVYVTHDQVEAMTMGDRVAVLCY
*: *** :* * : : .*:: ***** **: :.* :.::
Mflv_1083|M.gilvum_PYR-GCK GRLEQLAAPADLYAHPATPFVADFVG------LNNKVPASVS---GGTAT
Mvan_5739|M.vanbaalenii_PYR-1 GRLEQLAAPADLYANPATPFVAEFVG------LNNKVPATVS---GGSAS
MSMEG_6521|M.smegmatis_MC2_155 GKLEQLAAPAELYANPATPFVAEFVG------LHNKIPAEVT---DGKAR
TH_3667|M.thermoresistible__bu GRLAQVGAPADIYERPATPAAARFVG------ATVELPGQAR---DGVVH
Mb2419c|M.bovis_AF2122/97 GRIEQVGSPTDVYDAPANAFVMSFLG------AVSTLNGSLVRPHDIRVG
Rv2397c|M.tuberculosis_H37Rv GRIEQVGSPTDVYDAPANAFVMSFLG------AVSTLNGSLVRPHDIRVG
MAV_1785|M.avium_104 GRIEQIGSPTEVYDAPTNAFVMSFLG------AVSTLNGTLVRPHDIRVG
MMAR_3715|M.marinum_M GRIEQVGSPTEVYDAPANAFVMSFLG------AVSALNGALVRPHDIRVG
MUL_3658|M.ulcerans_Agy99 GRIEQVGSPTEVYDAPTNAFVMSFLG------AVSALNGALVRPHDIRVG
MAB_1655|M.abscessus_ATCC_1997 GRIEQVGSPEDLYDRPANSFVMSFLG------PVAKLNGILVRPHDIRVG
MLBr_01424|M.leprae_Br4923 GVLQQFATPRELYHKPENVFVAGFIGSPGMNLVTLSIVDSSVLLGDWPIR
* : *..:* ::* * . . *:* : .
Mflv_1083|M.gilvum_PYR-GCK -LLGVSVPALPGSVD-GDGVAMVRPEAVTVAADPAGT--ATVATVSFLGA
Mvan_5739|M.vanbaalenii_PYR-1 -LLGVSVAALPGSVD-GAGVAMVRPESVTVTADPAGA--STVSSVSFLGA
MSMEG_6521|M.smegmatis_MC2_155 -VLGATVPTLAGSVGSGAGTAMVRPESVTVQADPDGN--ATVASVAFLGP
TH_3667|M.thermoresistible__bu TALGSHPARLP--VPDGPVVVVLRPEELRLERSRDGVPAAAVQSVRFYGP
Mb2419c|M.bovis_AF2122/97 RTPNMAVAAADGTAGSTGVLRAVVDRVVVLGFEVRVELTSAATGGAFTAQ
Rv2397c|M.tuberculosis_H37Rv RTPNMAVAAADGTAGSTGVLRAVVDRVVVLGFEVRVELTSAATGGAFTAQ
MAV_1785|M.avium_104 RTPEMAVAAEDGTAESTGVARAIVDRVVKLGFEVRVELTSAATGGPFTAQ
MMAR_3715|M.marinum_M RTPEMAVAAADGTGESTGVTRAVVDRIVVLGFEVRVELTSAATGGAFTAQ
MUL_3658|M.ulcerans_Agy99 RTPEMAVAAADGTGESTGVTRAVVDRIVVLGFEVRVELTSAATGGAFTAQ
MAB_1655|M.abscessus_ATCC_1997 RNADLARAAAEGTGETTGVTKATVDRVVYLGFEVRVELTSAATGEQFTAQ
MLBr_01424|M.leprae_Br4923 IPREIASAASEVIIGVRPEHFELGSLGVEVEIDMVEELGADAYLYGRIAG
. : : . : . .
Mflv_1083|M.gilvum_PYR-GCK ITR---VGITMPDGSLLSAQMSSTAARALTPGDPVTVGVEPGGVLVTRPL
Mvan_5739|M.vanbaalenii_PYR-1 ISR---VHIMVPDGSTVSAQMASSAARGFAPGDPVTVGIEPRGVLVTRP-
MSMEG_6521|M.smegmatis_MC2_155 ISR---VYVNLPDGPVIHAQLPSSQARAFEAGQTVSVGLEPAPVLVV---
TH_3667|M.thermoresistible__bu QSA---VQVRLADDTVVTVLTAGDAG--VSVGEPVGVRVDGSVLAYPAPG
Mb2419c|M.bovis_AF2122/97 ITRGDAEALALREGDTVYVRATRVPPIAGG---VSGVDDAGVERVKVTST
Rv2397c|M.tuberculosis_H37Rv ITRGDAEALALREGDTVYVRATRVPPIAGG---VSGVDDAGVERVKVTST
MAV_1785|M.avium_104 ITRGDAEALALREGDTVYVRATRVPPITAGATTVPAVSRDGADEATLTSA
MMAR_3715|M.marinum_M ITRGDAEALALQEGDTVYVRATRVPPIAGA---APAVPDDGAGSTPSAAQ
MUL_3658|M.ulcerans_Agy99 ITRGDAEALALQEGDTVYVRATRVPPIAGA---APAVPDDGAGSTPRAAQ
MAB_1655|M.abscessus_ATCC_1997 ITRGDAEALGLREGDPVYVRVTRVP--------------DVGELAEVAS-
MLBr_01424|M.leprae_Br4923 ASKVTDQLVVARVDGRNPPAKGSRVRLYTEPGNVHFFGVDGHRIS-----
: : .
Mflv_1083|M.gilvum_PYR-GCK ------
Mvan_5739|M.vanbaalenii_PYR-1 ------
MSMEG_6521|M.smegmatis_MC2_155 ------
TH_3667|M.thermoresistible__bu RPPARS
Mb2419c|M.bovis_AF2122/97 ------
Rv2397c|M.tuberculosis_H37Rv ------
MAV_1785|M.avium_104 ------
MMAR_3715|M.marinum_M IGVGGQ
MUL_3658|M.ulcerans_Agy99 IGVGGQ
MAB_1655|M.abscessus_ATCC_1997 ------
MLBr_01424|M.leprae_Br4923 ------