For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTETQRPLDTEVIGDLASVIGRFRRQLRRATGGGFDAAGLTQSQGELLRLVGRKPGISVRESADELSLVP NTASTLVSRLVADGLLVKTADESDRRVGRLWLTPRAQRIADESRAARRAALSDVLAKLDDDQIEHLAKGL DVLAELTRLLQERDS
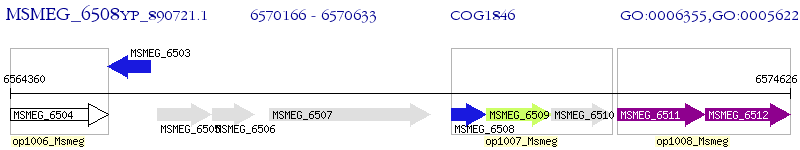
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6508 | - | - | 100% (155) | MarR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_3284 | - | 8e-11 | 32.59% (135) | transcriptional regulator, MarR family protein |
| M. smegmatis MC2 155 | MSMEG_6913 | - | 1e-09 | 32.19% (146) | putative transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0043c | - | 6e-09 | 28.97% (145) | MarR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_1034 | - | 3e-07 | 36.14% (83) | MarR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0042c | - | 6e-09 | 28.97% (145) | MarR family transcriptional regulator |
| M. leprae Br4923 | MLBr_02696 | - | 3e-08 | 29.20% (137) | putative MarR-family regulatory protein |
| M. abscessus ATCC 19977 | MAB_2472c | - | 4e-06 | 29.52% (105) | MarR family transcriptional regulator |
| M. marinum M | MMAR_0059 | - | 1e-07 | 30.82% (146) | transcriptional regulatory protein |
| M. avium 104 | MAV_5180 | - | 5e-40 | 62.22% (135) | MarR-family protein transcriptional regulator, putative |
| M. thermoresistible (build 8) | TH_0729 | - | 1e-05 | 34.67% (75) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0058 | - | 2e-07 | 30.14% (146) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_4913 | - | 1e-10 | 30.30% (132) | MarR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6508|M.smegmatis_MC2_155 --------------------------------------------------
MAV_5180|M.avium_104 --------------------------------------------------
Mb0043c|M.bovis_AF2122/97 ----MSVVRSIGKKMQRISGPNALAVKG----------------------
Rv0042c|M.tuberculosis_H37Rv ----MSVVRSIGKKMQRISGPNALAVKG----------------------
MMAR_0059|M.marinum_M --------------------------------------------------
MUL_0058|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02696|M.leprae_Br4923 MFSYITTKSWNPGNQSRYSRPRRLSAYNGLYPKYFDVLDFTNDDPLHPAP
TH_0729|M.thermoresistible__bu --------------------------------------------------
Mvan_4913|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1034|M.gilvum_PYR-GCK --------------------------------------------------
MAB_2472c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_6508|M.smegmatis_MC2_155 ----------------------------MTETQRPLDTEVIGDLASVIGR
MAV_5180|M.avium_104 ----------------------------------------------MVGR
Mb0043c|M.bovis_AF2122/97 ---RPTQVYGHT------HVRLDCRFMADSEFTAPEVTQLAEGLHRALSK
Rv0042c|M.tuberculosis_H37Rv ---RPTQVYGHT------HVRLDCRFMADSEFTAPEVTQLAEGLHRALSK
MMAR_0059|M.marinum_M --------------------------MADSESTAPEVTELAEGLHRALSK
MUL_0058|M.ulcerans_Agy99 --------------------------MADSESTAPEVTELAEGLHRALSK
MLBr_02696|M.leprae_Br4923 PHLRPPLQLAHTRSLNARHIALNCRLMADSEFTAHEVTELADGLHRALSK
TH_0729|M.thermoresistible__bu ----------------------------MKPPAVDPSTSLGADLLWVVSR
Mvan_4913|M.vanbaalenii_PYR-1 ---------------------------MGGRAASGASAQRAGRLADAFGR
Mflv_1034|M.gilvum_PYR-GCK -----------------------MCYAYFVRSRYDQLDDLLTRLHLARQR
MAB_2472c|M.abscessus_ATCC_199 -------------------------------------------MGILVSQ
:
MSMEG_6508|M.smegmatis_MC2_155 FRRQLRRATGGGFDAAGLTQSQGELLRLVGRKPGISVRESADELSLVPNT
MAV_5180|M.avium_104 FRRQLRRSAGRAFDPARLSESQSELLWLVGRRPGISVSAAAAELGLVPNT
Mb0043c|M.bovis_AF2122/97 LISMLRRGDPNGAAAGDLTLAQLSILVTLLDQGPIRMTDLAAHERVRTPT
Rv0042c|M.tuberculosis_H37Rv LISMLRRGDPNGAAAGDLTLAQLSILVTLLDQGPIRMTDLAAHERVRTPT
MMAR_0059|M.marinum_M LFSILRRGDPNGVVAGELTLAQLSILVTLLDRGPIRMTDLAAHERVRTPT
MUL_0058|M.ulcerans_Agy99 LFSILRRGDPNGVVAGELTLAQLSILVTLLDRGPIRMTDLAAHERVRTPT
MLBr_02696|M.leprae_Br4923 LFSILRRRTPNGAVSGELTLAQLSILVTLLDQGPIRMTDLAAHERVRTPT
TH_0729|M.thermoresistible__bu INRLATQR-----IRLPLPVAQARLLSTIQDRGSARISDLAVLDHCSQPT
Mvan_4913|M.vanbaalenii_PYR-1 AGKSVVRAFDDRLGEHGVSTPRSKLLAEVARMQPVRLADLAREVGISQGT
Mflv_1034|M.gilvum_PYR-GCK PQWRRRVLGPDSPVSGVSTIRVLRAVEQFDESGGASVRDVADFLAVEQST
MAB_2472c|M.abscessus_ATCC_199 MYRLLWSRASVIVRDFGVTVPQMECLAVLSAQPGISNVEIAAELRITPQA
. : . * :
MSMEG_6508|M.smegmatis_MC2_155 ASTLVSRLVADGLLVKTADESDRRVGRLWLTPRAQRIADESRAARRAALS
MAV_5180|M.avium_104 ASTLVTKLVSGGFLLRAAADTDRRVCQLRLTESAQQIVDESRAARRALLS
Mb0043c|M.bovis_AF2122/97 TTVAIRRLEKIGLVKRSRDPSDLRAVLVDITPQGRAVHGESLANRRAALA
Rv0042c|M.tuberculosis_H37Rv TTVAIRRLEKIGLVKRSRDPSDLRAVLVDITPQGRAVHGESLANRRAALA
MMAR_0059|M.marinum_M TTVAIRRLEKIGLVKRSRDPSDLRAVLVDITPQGRSVHAESLANRHAALA
MUL_0058|M.ulcerans_Agy99 TTVAIRRLEKIGLVKRSRDPSDLRAVLVDITPQGRSVHAKSLANRHAALA
MLBr_02696|M.leprae_Br4923 TTVAIRRLEKIGLVKRSRDLSDLRAVLVDITPRGRTVHGESLANRRAALT
TH_0729|M.thermoresistible__bu MTTQVRRLEEAGLVSRTVDPDDGRAVRISITPEGVRTLAQVRADRAAAID
Mvan_4913|M.vanbaalenii_PYR-1 ASTLVEALVRDGLVARGVDDNDRRAIRLTTTPEGETQARNWLRDYVVVAG
Mflv_1034|M.gilvum_PYR-GCK ASRLVAPAVAAGLLTKSSAADDQRRRTLVLTEVGRKELAEIAERRRELVA
MAB_2472c|M.abscessus_ATCC_199 VSLVQRSLEEAGLVHRSRRQADARVLTTELTAKGRKLFERADATLREDDE
: *:: : * * * . .
MSMEG_6508|M.smegmatis_MC2_155 DVLAKLDDDQIEHLAKGLDVLAELTRLLQERDS----------------
MAV_5180|M.avium_104 EVVDELNDDQIEALTRGLEVLDMMTQKLRERRP----------------
Mb0043c|M.bovis_AF2122/97 ALLSQLPRSDLETLRKALAPLERLASGEPASGPASNSPARKRA------
Rv0042c|M.tuberculosis_H37Rv ALLSQLPRSDLETLRKALAPLERLASGEPASGPASNSPARKRA------
MMAR_0059|M.marinum_M AMLDQLPHSDLETLMTALTPLERLACGEPANAPEDDAATPGRG------
MUL_0058|M.ulcerans_Agy99 AMLDQLPHSDLETLMTALTPLERLACGEPANAPEDDAATPGRG------
MLBr_02696|M.leprae_Br4923 TMLSQLPTSDLNILKNALAPLQRLASGEPVSSPAGDSPACKQA------
TH_0729|M.thermoresistible__bu PYIDRLSDTDRAVLADAVRVMRQLLADAADPAVLPADPPSRPHESTELR
Mvan_4913|M.vanbaalenii_PYR-1 EIFSCLTAEEQRQLTRLLDRIAESLDSPA--------------------
Mflv_1034|M.gilvum_PYR-GCK ETVADLPDSDVDTLVELLGRITDRFERAAR-------------------
MAB_2472c|M.abscessus_ATCC_199 EILANMTERDRVQLRRLLGGVIETISAR---------------------
. : : * : :