For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSDDVPLLRNRSGTARERDPQAPVEELEFEAAIGRNVRQLRQQHGLTVAEMAARVGISKAMMSKIENAQT SCSLSTLALLAKGLDVPVTSLFRGADVERPAAFVKAGTGPEIVRNGTKQGHEYQLLSSLRGEHKRLECLH VTLTEKSQTYPLFQHPGTEFIYMLEGVMDYSHSRSVYRLQPGDSLQIDGEGAHGPVDLVELPIRFLSVIA FPDSQV
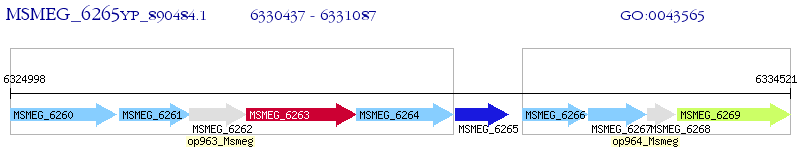
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6265 | - | - | 100% (216) | transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_4304 | - | 2e-10 | 25.53% (188) | regulatory protein |
| M. smegmatis MC2 155 | MSMEG_1151 | - | 8e-10 | 28.65% (178) | DNA-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1295 | - | 1e-108 | 90.65% (214) | XRE family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4363c | - | 3e-09 | 28.48% (165) | putative DNA-binding protein |
| M. marinum M | MMAR_2152 | - | 3e-11 | 27.22% (158) | transcriptional regulator |
| M. avium 104 | MAV_3453 | - | 9e-15 | 31.65% (158) | transcriptional regulator |
| M. thermoresistible (build 8) | TH_0145 | - | 3e-10 | 25.67% (187) | regulatory protein |
| M. ulcerans Agy99 | MUL_1737 | - | 2e-11 | 27.22% (158) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_5514 | - | 1e-105 | 88.84% (215) | XRE family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1295|M.gilvum_PYR-GCK MANQNDRDDHTPLLRNTSGTARDRDPGAPVEELEIESAIGRNVRLLRQQQ
Mvan_5514|M.vanbaalenii_PYR-1 MALPEDPADPGPLLRNTSGTARDRDPAAPVEELEIEAAIGRNVRLLRLQQ
MSMEG_6265|M.smegmatis_MC2_155 MS------DDVPLLRNRSGTARERDPQAPVEELEFEAAIGRNVRQLRQQH
MMAR_2152|M.marinum_M --------------------------------------MTALLRAVRRQR
MUL_1737|M.ulcerans_Agy99 --------------------------------------MTALLRAVRRQR
MAV_3453|M.avium_104 --------------------------------------MTALLRPVRRQR
TH_0145|M.thermoresistible__bu -------------------------MEANQREGRIEDRIRQRLRELRKQR
MAB_4363c|M.abscessus_ATCC_199 -------------------------------MTDHNTLVARNVRRYRQER
: :* * ::
Mflv_1295|M.gilvum_PYR-GCK GLTVAETAARVGISKAMMSKIENAQTSCSLSTLALLAKGFDVPVTSLFRG
Mvan_5514|M.vanbaalenii_PYR-1 GLTVAETAARVGISKAMMSKIENAQTSCSLSTLALLAKGFDVPVTSLFRG
MSMEG_6265|M.smegmatis_MC2_155 GLTVAEMAARVGISKAMMSKIENAQTSCSLSTLALLAKGLDVPVTSLFRG
MMAR_2152|M.marinum_M GLTLEQLAQRAGLTKSYLSKIERGQSTPSIAVALKVARALDVDVGRLFSD
MUL_1737|M.ulcerans_Agy99 GLTLEQLAQRAGLTKSYLSKIERGQSTPSIAVALKVARALDVDVGRLFSD
MAV_3453|M.avium_104 GLTLEALAAQTGLTKSYLSKIERGQSTPSIAVALKVARALDVDVGRLFSD
TH_0145|M.thermoresistible__bu GLTLEDVATRAQIDVSTLSRLESGKRRLAIDHLPRLAAALSVSTDDLLRA
MAB_4363c|M.abscessus_ATCC_199 ALSLGDLARRSGLSKQTVSKIEQGVGNPTVETLALLGEALQVPPQRLLTE
.*:: * : : :*::* . :: :. .:.* *:
Mflv_1295|M.gilvum_PYR-GCK ADVERPAAFVKAGTGARIVREGTREGHEYQLLGSLRGEHKRLECLEVTLS
Mvan_5514|M.vanbaalenii_PYR-1 ADVERPAAFVKAGTGARIVREGTREGHEYQLLGSLRGEHKRLECLEVTLS
MSMEG_6265|M.smegmatis_MC2_155 ADVERPAAFVKAGTGPEIVRNGTKQGHEYQLLSSLRGEHKRLECLHVTLT
MMAR_2152|M.marinum_M ESAHETITVDRAHD-----RLGPQSGRYHVLASTQLGKTMSPFVVRPTAG
MUL_1737|M.ulcerans_Agy99 ESAHETITADRAHD-----RLGPQSGRYHVLASTQLGKTMSPFVVRPTAG
MAV_3453|M.avium_104 EAAREKIAVERAG--------GPGGRRYRVLASSMLGKTMSPFVIRPT--
TH_0145|M.thermoresistible__bu PDTEDPRVRGSAHT--------SHGVTYWPLTRQGPAGGLHAFKIRISAR
MAB_4363c|M.abscessus_ATCC_199 WGTPVFVQRASEDG-----WVDKGARSERLLDEIYGSGYVRNLVVRLERT
. * . :.
Mflv_1295|M.gilvum_PYR-GCK -EKSQTYPLFQHPGTEFIYMLEGVMDYSHSRSVYRLQPGDSLQIDGEGAH
Mvan_5514|M.vanbaalenii_PYR-1 -GKSQTYPLFQHPGTEFIYMLEGVMDYSHSRTVYRLHPGDSLQIDGEGAH
MSMEG_6265|M.smegmatis_MC2_155 -EKSQTYPLFQHPGTEFIYMLEGVMDYSHSRSVYRLQPGDSLQIDGEGAH
MMAR_2152|M.marinum_M QDCASDGPHPEHTGQEFVFVHTGSVELDYGDQTITLGTGDSAYFDASVGH
MUL_1737|M.ulcerans_Agy99 QDCASDGPHPEHTGQEFVFVHTGSVELDYGDQTITLGTGDSAYFDASVGH
MAV_3453|M.avium_104 -EQLADDPHPEHAGQEFLFVHAGRVELDYGDQTFTLGPGDSAYFDASVSH
TH_0145|M.thermoresistible__bu -RRTPPAGLPVHEGHDWVYVLSGHMRLILDSRDFVIKPGEAVEFSTWTPH
MAB_4363c|M.abscessus_ATCC_199 AQAGEEIGAHAPGTLHHVYVIEGILRTGPLNDPVELRTGDFARFPGDVPH
. ::: * : : .*: : *
Mflv_1295|M.gilvum_PYR-GCK GPVDLVEVPIRFLSVIAFPDSQV----
Mvan_5514|M.vanbaalenii_PYR-1 GPVDLVEVPIRFLSVIAFPDSQV----
MSMEG_6265|M.smegmatis_MC2_155 GPVDLVELPIRFLSVIAFPDSQV----
MMAR_2152|M.marinum_M KLRAVGTERAEVVVVAGAELGGR----
MUL_1737|M.ulcerans_Agy99 KLRAVGTERAEVVVVAGAELGGR----
MAV_3453|M.avium_104 KVRSVGPEPAEVVAVAHAEPGSAAFAG
TH_0145|M.thermoresistible__bu WFGAVDG-PVEAIMILGPHGERLHLHE
MAB_4363c|M.abscessus_ATCC_199 RHESISPRVVAHLVTTLPQIRQFDPIR
: :