For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KIVAADVLVTAPGRNYVTLKITTDDGLVGWGDATLNGRELAVASYLRDHVCPQLIGRDAQRIEDTWQYFY KGAYWRRGPVTMTAIAAVDIALWDIKGKAAGMPLYQLLGGAARDGVMVYGHASGASLEEMLDDFAGYVDR GYRAVRAQCAVPGMGKVYGVHPGSSLYEPASAHLPAEESWETGPYLRFAPRMLGAVRERFGFDVHLLHDV HHRLSPIEAARLGRDVEEHRLFWMEDPTPAEDQEAFRLIRQNTTTPLAVGEVFNSIWDCQHLITERLIDY IRTSVSHAGGVTHLRRIFALAELYGVRTGSHGAGDLSPVSFAAALHIDMSVPNFGVQEYMGHVEPADEVF RTNWTFQNGLMHPGDAPGLGVEVDEQAAARFPYDPKSLPVNRRADGSVHDW*
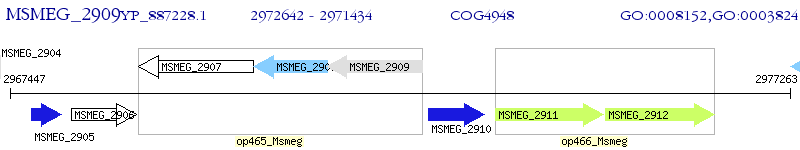
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2909 | - | - | 100% (402) | starvation-sensing protein RspA |
| M. smegmatis MC2 155 | MSMEG_6177 | - | 3e-51 | 31.63% (411) | galactonate dehydratase |
| M. smegmatis MC2 155 | MSMEG_0126 | - | 1e-25 | 26.23% (366) | mandelate racemase/muconate lactonizing enzyme |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1357 | - | 2e-12 | 23.04% (369) | mandelate racemase/muconate lactonizing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3096 | - | 5e-20 | 24.80% (375) | mandelate racemase/muconate lactonizing enzyme |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1029 | - | 5e-09 | 20.70% (372) | mandelate racemase/muconate lactonizing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1357|M.gilvum_PYR-GCK ---MKITAVDAIPFAIPYTKPLKFASGEVHTAEHVLVRVHTDEGVVGVAE
Mvan_1029|M.vanbaalenii_PYR-1 MAPIRVTGARITPVAFADPPLLNTVGVHQPYALRAIIQLDTDAGLVGLGE
MAV_3096|M.avium_104 --------MTPDPAIDEVTARVYQIPTDAPEADGTLSWSSTTLVLAEVAA
MSMEG_2909|M.smegmatis_MC2_155 -----------------MKIVAADVLVTAPGRNYVTLKITTDDGLVGWGD
. * :. .
Mflv_1357|M.gilvum_PYR-GCK APPRPFTYGETQDGILAVIR-QIFAPQIVGLTLTEREVVNARLARTVGNP
Mvan_1029|M.vanbaalenii_PYR-1 TYADTRHLARLRAAAEAITGLDVFALNRIRASIGSRLEGDTTAVGTAGMI
MAV_3096|M.avium_104 GGRRGIGYTYADGACAALIT-GALAGSVTGHSALDVPARWQAMVRAMRNN
MSMEG_2909|M.smegmatis_MC2_155 ATLNGR-----ELAVASYLR-DHVCPQLIGRDAQRIEDTWQYFYKGAYWR
. : .. .
Mflv_1357|M.gilvum_PYR-GCK TAKAAVD-------MAIWDALGRSLDLSVSALLGGYG-DHMRVSHML-GF
Mvan_1029|M.vanbaalenii_PYR-1 TSASVVDQVLSPFEVACLDVQGQSLGRPVSDLLGGAVRDAVPFSAYL-FY
MAV_3096|M.avium_104 GRPGLVSCAISAVDTALWDLKGKLLGLPVCRLLGMAH-DSVPIYGSG-GF
MSMEG_2909|M.smegmatis_MC2_155 RGP-VTMTAIAAVDIALWDIKGKAAGMPLYQLLGGAARDGVMVYGHASGA
. * * *: . .: *** * : .
Mflv_1357|M.gilvum_PYR-GCK D----------------TPDTMVAEAERIRDEYGIS-----------TFK
Mvan_1029|M.vanbaalenii_PYR-1 KWAGHPNAEPDRFGEAMDPNGLVAQARRIIDEYGFT-----------AIK
MAV_3096|M.avium_104 TS--------------YDDNATRAQLERWVHEWRIP-----------RVK
MSMEG_2909|M.smegmatis_MC2_155 SLEEMLDDFAGYVDRGYRAVRAQCAVPGMGKVYGVHPGSSLYEPASAHLP
. . : . .
Mflv_1357|M.gilvum_PYR-GCK VKVGR---RPVTLDTAVVRALRERFGDTVELYVDGNRGWTASESLRALKD
Mvan_1029|M.vanbaalenii_PYR-1 LKGGV---FPPEEEMAAIEALSRNFPG-LPLRLDPNAAWTPHTAVKVASG
MAV_3096|M.avium_104 IKIGESWGSHERRDLDRIALARTVIGPDTELYVDANGGYRRKQAIRMAHA
MSMEG_2909|M.smegmatis_MC2_155 AEESWETGPYLRFAPRMLGAVRERFGFDVHLLHDVHHRLSPIEAARLGRD
: . : : * * : : :
Mflv_1357|M.gilvum_PYR-GCK MADLDLLFAEELCPADDVLGRRWLVEHTEIPFIADESVPTPADVTREVLG
Mvan_1029|M.vanbaalenii_PYR-1 LAGILEYLED---PTPGLAGMAEVAQQAPMPLATNMCVVAFDQLAPAVTK
MAV_3096|M.avium_104 MAGHDVTWFEEPVSSDDLAGLREVRDQVSADVTAGEYGYDLAYFHRMLAA
MSMEG_2909|M.smegmatis_MC2_155 VEEHRLFWMEDPTPAEDQEAFRLIRQNTTTPLAVGEVFNSIWDCQHLITE
: : .: . . : ::. . .. :
Mflv_1357|M.gilvum_PYR-GCK -GSATAISIKTARTGFTGSRRVHHLAEGLGLEVVMGNQIDGQLGTACAVA
Mvan_1029|M.vanbaalenii_PYR-1 NAVRVVLSDHHYWGGLQRSRLLAGICDTFGLGLSMHSNSHLGISLAAMVH
MAV_3096|M.avium_104 DAVDCLQIDVTRCGGITDWLRAAAVAAAHNVDVSGHCAPNLHAHVATAVA
MSMEG_2909|M.smegmatis_MC2_155 RLIDYIRTSVSHAGGVTHLRRIFALAELYGVRTGSHGAGDLSP-VSFAAA
*. :. .: . : .
Mflv_1357|M.gilvum_PYR-GCK FGAAFELTSRRAGELSNFLDMSDDLLTHPLRIRDGRLHVPPG-PGLGIEI
Mvan_1029|M.vanbaalenii_PYR-1 LAGATPNLT-YACDTHWPWKTEDVVKDGALAFVDGAVPVPTS-PGLGVEI
MAV_3096|M.avium_104 N-------LRHLEYFHDHHRIERMFFDGALSPQGGVLRPDQQRPGFGLEF
MSMEG_2909|M.smegmatis_MC2_155 LHIDMSVPNFGVQEYMGHVEPADEVFRTNWTFQNGLMHPGDA-PGLGVEV
. .* : **:*:*.
Mflv_1357|M.gilvum_PYR-GCK DPDKLDHYRTDH----------------------------
Mvan_1029|M.vanbaalenii_PYR-1 DDDALDALHEQYVRCGIRDRDDTGYMRTVDPSFEPAGPRW
MAV_3096|M.avium_104 KQRDADRYRMR-----------------------------
MSMEG_2909|M.smegmatis_MC2_155 DEQAAARFPYDPKSLPVNRRADGSVHDW------------
.