For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TAEVRLHRISVVMSTMRRNRPVVDGVSLRVAPGEMVAVVGPNGAGKTTLLRTIYRAQRPTAGNVLIDGAD LWKLPRKRAAQRVAAVLQEMTTDFELTVYDIVAMGRTPYKRSFESDNATDREIIASALAALDIADLAHAP FGRLSGGQKQRVLIARSLAQHTDVIVLDEPTNHLDLRHQHDTLRLLRATGSTVIAALHDLNLAAAYCDRI CVLDRGQVVAVGAPREVLTVELLAEVYGVDATIGDTAGVPQITVVPGQAAPR*
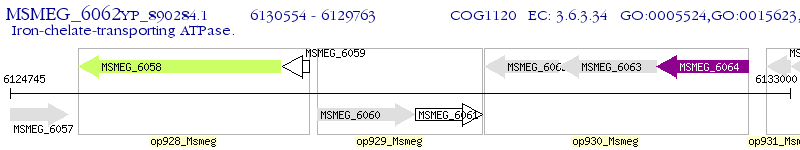
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6062 | - | - | 100% (263) | Fe uptake system permease |
| M. smegmatis MC2 155 | MSMEG_4557 | - | 5e-52 | 44.63% (242) | ABC transporter, ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_0015 | - | 2e-43 | 38.02% (242) | ferric enterobactin transport ATP-binding protein FepC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2419c | cysA1 | 4e-24 | 33.49% (209) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. gilvum PYR-GCK | Mflv_3884 | - | 2e-61 | 52.68% (224) | ABC transporter related |
| M. tuberculosis H37Rv | Rv2397c | cysA1 | 4e-24 | 33.49% (209) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. leprae Br4923 | MLBr_00669 | ftsE | 1e-24 | 32.04% (206) | putative cell division ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_0548c | - | 4e-58 | 47.64% (233) | putative iron compound ABC transporter, ATP-binding protein |
| M. marinum M | MMAR_3715 | cysA1 | 8e-26 | 35.41% (209) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. avium 104 | MAV_0058 | - | 3e-26 | 35.41% (209) | ABC transporter, ATP-binding protein |
| M. thermoresistible (build 8) | TH_3040 | - | 6e-57 | 48.46% (227) | PUTATIVE putative ABC transporter, ATP-binding protein |
| M. ulcerans Agy99 | MUL_3658 | cysA1 | 7e-26 | 35.41% (209) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. vanbaalenii PYR-1 | Mvan_6048 | - | 2e-27 | 36.84% (209) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3884|M.gilvum_PYR-GCK ----------MTAARLSTAGLCVDLGGRR----IVEDVSMDVPAGRVVGI
MAB_0548c|M.abscessus_ATCC_199 -------------MDVTFRNVSVSIAGAQ----IISDIHLNVAAGGFVGI
TH_3040|M.thermoresistible__bu -MTDVRDATATAQTAVCADRVCWTVRGTL----IVDGVSLDVTPGSTVGL
MSMEG_6062|M.smegmatis_MC2_155 -----------MTAEVRLHRISVVMSTMRRNRPVVDGVSLRVAPGEMVAV
Mb2419c|M.bovis_AF2122/97 ------MT-------YAIVVADATKRYG--DFVALDHVDFVVPTGSLTAL
Rv2397c|M.tuberculosis_H37Rv ------MT-------YAIVVADATKRYG--DFVALDHVDFVVPTGSLTAL
MMAR_3715|M.marinum_M MKKETSLTGPGT-GGHAIVVRDAFKQYG--DFVALDHVDFVVPAGSLTAL
MUL_3658|M.ulcerans_Agy99 ------MTGPGT-GGHAIVVRDAFKQYG--DFVALDHVDFVVPAGSLTAL
MAV_0058|M.avium_104 ------MSNPASRASVSLACKDIHQTLN--GNAVLRGVDLDTPAGSTVAV
Mvan_6048|M.vanbaalenii_PYR-1 ------MT-----TAVSLTAKDIHLSFG--KSAVLRGVDLDVAAGSSTAV
MLBr_00669|M.leprae_Br4923 ----------------MITLDHVSKKYKSLARPALDNVNVKIDKGEFVFL
: : . * . :
Mflv_3884|M.gilvum_PYR-GCK VGPNGCGKSTLLRTLGGILRPASGVVSIDGTDIATLGARQL---ARTVAA
MAB_0548c|M.abscessus_ATCC_199 VGPNGSGKSTTLRCLYRALTPASGDVAVNGVSVRAISMREN---ARQVAA
TH_3040|M.thermoresistible__bu LGPNGSGKSTLLRMLAGIRRPSSGVIRLHDQDVTTLARRQV---ARRVAV
MSMEG_6062|M.smegmatis_MC2_155 VGPNGAGKTTLLRTIYRAQRPTAGNVLIDGADLWKLPRKRA---AQRVAA
Mb2419c|M.bovis_AF2122/97 LGPSGSGKSTLLRTIAGLDQPDTGTITINGRDVTRVPPQ-----RRGIGF
Rv2397c|M.tuberculosis_H37Rv LGPSGSGKSTLLRTIAGLDQPDTGTITINGRDVTRVPPQ-----RRGIGF
MMAR_3715|M.marinum_M LGPSGSGKSTLLRTIAGLDQPDSGNITINGRDVTRVPPQ-----RRGIGF
MUL_3658|M.ulcerans_Agy99 LGPSGSGKSTLLRTIAGLDQPDSGNITINGRDVTRVPPQ-----RRGIGF
MAV_0058|M.avium_104 IGPSGSGKSTLLRTLNRLYEPDRGDILLDGRSVLHDDPDQL---RQRIGM
Mvan_6048|M.vanbaalenii_PYR-1 IGPSGSGKSTLLRTLNRLYEPDRGDILLDGRSVLGDNPDQL---RQRIGM
MLBr_00669|M.leprae_Br4923 IGPSGSGKSTFMRLLLGAETPTSGDVRVSKFHVNKLPGRHIPRLRQVIGC
:**.*.**:* :* : * * : : : : :.
Mflv_3884|M.gilvum_PYR-GCK VLQDSAGDFDLRVRDVVLMGRSPYKRMFEGDNASDIRIVGDSLALVDAAH
MAB_0548c|M.abscessus_ATCC_199 LTQDMSLDFDFTAAEVIATGRLPHGASLRGTSQLDRQICARATETADVTA
TH_3040|M.thermoresistible__bu VEQESGTDSDPRVRDVIALGRIPHRRPWAALSDRDLRIIERAAAATDVAD
MSMEG_6062|M.smegmatis_MC2_155 VLQEMTTDFELTVYDIVAMGRTPYKRSFESDNATDREIIASALAALDIAD
Mb2419c|M.bovis_AF2122/97 VFQHYAAFKHLTVRDNVAFG----LKIRKRPKAEIKAKVDNLLQVVGLSG
Rv2397c|M.tuberculosis_H37Rv VFQHYAAFKHLTVRDNVAFG----LKIRKRPKAEIKAKVDNLLQVVGLSG
MMAR_3715|M.marinum_M VFQHYAAFKHLTVRDNVAYG----LKIRKRPKAEIRDKVDNLLEVVGLSG
MUL_3658|M.ulcerans_Agy99 VFQHYAAFKHLTVRDNVAYG----LKIRKRPKAEIRDKVDNLLEVVGLSG
MAV_0058|M.avium_104 VFQHFNLFPHRTVLDNVALAP---RKLRRLPAEAARELALTQLDRVGLKH
Mvan_6048|M.vanbaalenii_PYR-1 VFQHFQLFPHRTVLDNVTLAP---RKLKRLSGDEARELGMAQLDRVGLRN
MLBr_00669|M.leprae_Br4923 VFQDFRLLQQKTVYENVAFAL---EVIGRRS-DAINQVVPDVLETVGLSG
: *. . . : : . .
Mflv_3884|M.gilvum_PYR-GCK LEDRPFPLLSGGERRRVLVARALAQQPHLLLMDEPTNHLDVRHTFDVLAL
MAB_0548c|M.abscessus_ATCC_199 LSGRTFGSLSGGERQRVLIARALAQQPHVLVLDEPTNHLDVRHQYSILNS
TH_3040|M.thermoresistible__bu RLDQHYATLSGGERQRVQLSRAFAQEPEVLLLDEPTNHLDVRHQLDLMRM
MSMEG_6062|M.smegmatis_MC2_155 LAHAPFGRLSGGQKQRVLIARSLAQHTDVIVLDEPTNHLDLRHQHDTLRL
Mb2419c|M.bovis_AF2122/97 FQSRYPNQLSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREELRAW
Rv2397c|M.tuberculosis_H37Rv FQSRYPNQLSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREELRAW
MMAR_3715|M.marinum_M FHGRYPNQLSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREDLRAW
MUL_3658|M.ulcerans_Agy99 FHGRYPDQLSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREDLRAW
MAV_0058|M.avium_104 KAGARPATLSGGQQQRVAIARALAMSPQVMFFDEATSALDPEMVKGILQL
Mvan_6048|M.vanbaalenii_PYR-1 KADARPATLSGGQQQRVAIARALAMSPQVMFFDEATSALDPELVKGVLEL
MLBr_00669|M.leprae_Br4923 KANRLPDELSGGEQQRVAIARAFVNRPLVLLADEPTGNLDPDTSKDIMDL
****:::*: ::*::. . ::. **. . **
Mflv_3884|M.gilvum_PYR-GCK PA----TLGVTAVIALHDLNLAAQYCDVIHVLRNGRQVAAGSPADVLTAD
MAB_0548c|M.abscessus_ATCC_199 AR----ALGITVIAALHDLNLAAQYCDTIHVMADGNVVLSGSPHEVLTAE
TH_3040|M.thermoresistible__bu VR----DAELTSVIAMHDLNLAAAFCDALLVLAHGRVVAAGPPAGVLTAE
MSMEG_6062|M.smegmatis_MC2_155 LR----ATGSTVIAALHDLNLAAAYCDRICVLDRGQVVAVGAPREVLTVE
Mb2419c|M.bovis_AF2122/97 LRRLHDEVHVTTVLVTHDQAEALDVADRIAVLHKGRIEQVGSPTDVYDAP
Rv2397c|M.tuberculosis_H37Rv LRRLHDEVHVTTVLVTHDQAEALDVADRIAVLHKGRIEQVGSPTDVYDAP
MMAR_3715|M.marinum_M LRRLHDEVHVTTVLVTHDQAEALDVADRIAVLNSGRIEQVGSPTEVYDAP
MUL_3658|M.ulcerans_Agy99 LRRLHDEVHVTTVLVTHDQAEALDVADRIAVLNSGRIEQVGSPTEVYDAP
MAV_0058|M.avium_104 IADLGAEG-MTMVVVTHEMGFARSASDAVIFMDHGKVVESGPPEQIFEAA
Mvan_6048|M.vanbaalenii_PYR-1 IADLGAEG-MTMVVVTHEMGFARSSADTVVFMDHGQVVETGPPEQLFEAA
MLBr_00669|M.leprae_Br4923 LERINRTG-TTVLMATHDHHIVDSMRQRVVELSLGRLVR-DEWCGIYGMD
* : . *: . : : : *. . :
Mflv_3884|M.gilvum_PYR-GCK LLGDVWEVRATVQPHPDTGRPHIYFDPRRRVTEESRSG------------
MAB_0548c|M.abscessus_ATCC_199 SVGRWFGIDCHVITHPRSGVPQVIFYAEPERQEKQ---------------
TH_3040|M.thermoresistible__bu LVERVYGVRARIG-HDDTG-LYVRFLG-----------------------
MSMEG_6062|M.smegmatis_MC2_155 LLAEVYGVDATIG--DTAGVPQITVVPGQAAPR-----------------
Mb2419c|M.bovis_AF2122/97 ANAFVMSFLGAVSTLNGSLVRPHDIRVGRTPNMAVAAADGTAGSTGVLRA
Rv2397c|M.tuberculosis_H37Rv ANAFVMSFLGAVSTLNGSLVRPHDIRVGRTPNMAVAAADGTAGSTGVLRA
MMAR_3715|M.marinum_M ANAFVMSFLGAVSALNGALVRPHDIRVGRTPEMAVAAADGTGESTGVTRA
MUL_3658|M.ulcerans_Agy99 TNAFVMSFLGAVSALNGALVRPHDIRVGRTPEMAVAAADGTGESTGVTRA
MAV_0058|M.avium_104 ETERLQRFLSQVL-------------------------------------
Mvan_6048|M.vanbaalenii_PYR-1 ETDRLRRFLSQVL-------------------------------------
MLBr_00669|M.leprae_Br4923 R-------------------------------------------------
Mflv_3884|M.gilvum_PYR-GCK --------------------------------------------------
MAB_0548c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3040|M.thermoresistible__bu --------------------------------------------------
MSMEG_6062|M.smegmatis_MC2_155 --------------------------------------------------
Mb2419c|M.bovis_AF2122/97 VVDRVVVLGFEVRVELTSAATGGAFTAQITRGDAEALALREGDTVYVRAT
Rv2397c|M.tuberculosis_H37Rv VVDRVVVLGFEVRVELTSAATGGAFTAQITRGDAEALALREGDTVYVRAT
MMAR_3715|M.marinum_M VVDRIVVLGFEVRVELTSAATGGAFTAQITRGDAEALALQEGDTVYVRAT
MUL_3658|M.ulcerans_Agy99 VVDRIVVLGFEVRVELTSAATGGAFTAQITRGDAEALALQEGDTVYVRAT
MAV_0058|M.avium_104 --------------------------------------------------
Mvan_6048|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mflv_3884|M.gilvum_PYR-GCK -------------------------------
MAB_0548c|M.abscessus_ATCC_199 -------------------------------
TH_3040|M.thermoresistible__bu -------------------------------
MSMEG_6062|M.smegmatis_MC2_155 -------------------------------
Mb2419c|M.bovis_AF2122/97 RVPPIAGGVSGVDDAGVERVKVTST------
Rv2397c|M.tuberculosis_H37Rv RVPPIAGGVSGVDDAGVERVKVTST------
MMAR_3715|M.marinum_M RVPPIAGAAPAVPDDGAGSTPSAAQIGVGGQ
MUL_3658|M.ulcerans_Agy99 RVPPIAGAAPAVPDDGAGSTPRAAQIGVGGQ
MAV_0058|M.avium_104 -------------------------------
Mvan_6048|M.vanbaalenii_PYR-1 -------------------------------
MLBr_00669|M.leprae_Br4923 -------------------------------