For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VALAAGRGAGVLRSLIRGSVRFVSYREQAPPPPLRDLVECSWERSGPATPARVLPDGCMDLIRLDGRVVV AGPDTTAYLAAASPDPVRGLRFRPGALPRVLGVPAAELRDTRVDLHDLRTVPSCDSLETLALTLASDLAP PRSETAPWSLAALRHVTRRFADGAAVDDVADDLGWSDRTLHRHCTSVYGYGPAMLRRVLRFRRAVRLLGS GTGLSDVAAQSGYADQPHLHREVRALARVGVRELLGQVSSAANRSTDVPSGSSTVA
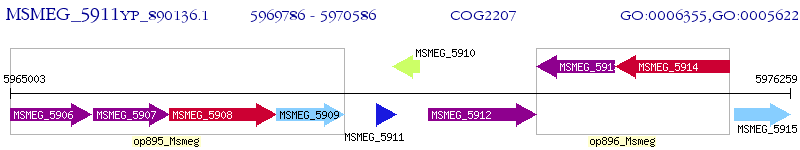
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5911 | - | - | 100% (266) | AraC family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_2590 | - | 5e-05 | 29.35% (201) | AraC-family protein transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3701c | - | 6e-09 | 35.16% (91) | AraC family transcriptional regulator |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_0221 | - | 4e-08 | 29.15% (247) | transcriptional regulator, AraC family protein |
| M. thermoresistible (build 8) | TH_2588 | - | 2e-07 | 28.48% (165) | PUTATIVE putative transcriptional regulator, AraC family |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_3701c|M.abscessus_ATCC_199 MRSPARYEGRTTADASWRAVSAAADVTLSGIAHGYTELR-ERAHRPVERS
TH_2588|M.thermoresistible__bu VRDPARIE-----------IGGHPAAPLRPFVTGYTGFD-LRGFEPGVHV
MAV_0221|M.avium_104 --------------------MYRPSPPLAEHIEYFGYWRGDEALGVHTSR
MSMEG_5911|M.smegmatis_MC2_155 ----------------MALAAGRGAGVLRSLIRGSVRFVSYREQAPPPP-
* .
MAB_3701c|M.abscessus_ATCC_199 EVAGVAPVLVVELDAPLLVADVADHA-SPR-IWQAFVAGVSQGPSSTVHS
TH_2588|M.thermoresistible__bu GMPSHTLTAVLTFGEPLEIDVPSGLS-GTSGRYPSMASGLLAQSISIRHR
MAV_0221|M.avium_104 ALPRGAVTAIIDVGGRTDLGFYASDARTPLTVPPAFAAGAGATAYVVRVA
MSMEG_5911|M.smegmatis_MC2_155 -LRDLVECSWERSGPATPARVLPDGCMDLIRLDGRVVVAGPDTTAYLAAA
: . . .. . .. . .
MAB_3701c|M.abscessus_ATCC_199 GAQHCIEVRLTPLGLHRVSGLAMDAVSNRVVSLEELFGKKARYLPERLAA
TH_2588|M.thermoresistible__bu GAQRGVKLSFTPLGARMVFGMPAGALANSVVPLSDVLADLWPELEDRLGA
MAV_0221|M.avium_104 PAHTVMTIHFRPAGALAFLGSPLSDLEDALVGLEDLWGRDAALLREQLID
MSMEG_5911|M.smegmatis_MC2_155 SPDPVRGLRFRPGALPRVLGVPAAELRDTRVDLHDLRTVPSCDSLETLAL
.. : : * . . * . : : * * :: : *
MAB_3701c|M.abscessus_ATCC_199 EPNWVRRFDLLDSVFMRAAAE-----GPEADAEVEFAWHRLNRTGGTAGI
TH_2588|M.thermoresistible__bu AATWPHRFRVLDDVLARAVARTMCAGMFDVRPEVAAAWQRLVDSRGQVRV
MAV_0221|M.avium_104 AGSPPRRVALLQAFLVRRMRR-----NAVWPPARLAPVLRGADLDPSMRV
MSMEG_5911|M.smegmatis_MC2_155 T---------LASDLAPPRSE-------TAPWSLAALRHVTRRFADGAAV
* : . :
MAB_3701c|M.abscessus_ATCC_199 GEILNEIGWSRARLAQRFRSQVGLTPKAAARVLRFNRAMTLLNQPGHRSL
TH_2588|M.thermoresistible__bu GDLAHDLGCSRRHLTARFREEFGLTPKTLARILRLEQAVGLVAAAGAPAW
MAV_0221|M.avium_104 SKAQELSGLSRKRFAALFRCEVGLSPKAYLRVLRLQAALRALDTPARG--
MSMEG_5911|M.smegmatis_MC2_155 DDVADDLGWSDRTLHRHCTSVYGYGPAMLRRVLRFRRAVRLLG--SGTGL
.. . * * : * * *:**:. *: : .
MAB_3701c|M.abscessus_ATCC_199 SSIALACGYFDQAHFNRDFRVLAGCSPRQWAALRHEDLMGYRIPADGEHL
TH_2588|M.thermoresistible__bu AEVSAAAGYADQAHLVRDWRSFTGRTPSSWFDG-----------------
MAV_0221|M.avium_104 ATIAADLGYFDQAHFVREFRAFTGATPTQYARR-----------------
MSMEG_5911|M.smegmatis_MC2_155 SDVAAQSGYADQPHLHREVRALARVGVRELLGQVSSAAN-----------
: :: ** **.*: *: * :: .
MAB_3701c|M.abscessus_ATCC_199 YKTGVDGGPSVGTVL
TH_2588|M.thermoresistible__bu -DVLVDGRPHG----
MAV_0221|M.avium_104 -RSSMPGHVELAR--
MSMEG_5911|M.smegmatis_MC2_155 RSTDVPSGSSTVA--
: .