For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSLAATSSGICVSFGTNLRKDQQMTCEHAPLTTRRLPFGVAVLGGPTTVVDIGGHRLICDPTFDPPTDYG YLQKLAGPAADESAVGDVDVALVSHDLHPDNLDVAGRAFALSAPVLLTTATAATRLGAPSVPLAPWTTWE SPDGLAITAVPARHGPADGELNEEGFVNCEVIGFVVTAPDSPTVYLSGDNASVEIVREIRERIGGIDCAV LFAGSASVPAKFDGRPLSLTAERAAAAAEVLGSPHVVVAHQDGWAHFRQGAADTAAAFHAAGIGDRLCSA PLGHMCAI
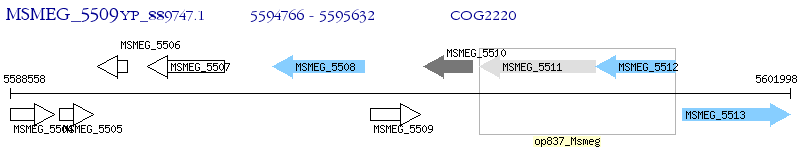
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5509 | - | - | 100% (288) | hypothetical protein MSMEG_5509 |
| M. smegmatis MC2 155 | MSMEG_6233 | - | 1e-15 | 25.74% (272) | hypothetical protein MSMEG_6233 |
| M. smegmatis MC2 155 | MSMEG_5109 | - | 5e-12 | 26.21% (248) | hypothetical protein MSMEG_5109 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0930 | - | 2e-06 | 30.40% (125) | hypothetical protein Mb0930 |
| M. gilvum PYR-GCK | Mflv_2585 | - | 4e-13 | 26.69% (266) | protein Pfam: Lactamase_B |
| M. tuberculosis H37Rv | Rv0906 | - | 2e-06 | 30.40% (125) | hypothetical protein Rv0906 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_1029 | - | 8e-07 | 32.00% (125) | metallo-beta-lactamase family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0930|M.bovis_AF2122/97 MVRRALRLAAGTASLAAGTWLLRALHGTPAALGADAASIRAVSEQSPNYR
Rv0906|M.tuberculosis_H37Rv MVRRALRLAAGTASLAAGTWLLRALHGTPAALGADAASIRAVSEQSPNYR
MAV_1029|M.avium_104 -MRGTLRLVAGTASLAAGGWLVRALHGAPDALGADPASIAAVTSGSPHFR
Mflv_2585|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5509|M.smegmatis_MC2_155 --------------------------------------------------
Mb0930|M.bovis_AF2122/97 DGAFVNLDPASMFTLDREELRLIVWELVARHSASRPAAPIPLASPNIYRG
Rv0906|M.tuberculosis_H37Rv DGAFVNLDPASMFTLDREELRLIVWELVARHSASRPAAPIPLASPNIYRG
MAV_1029|M.avium_104 DGAFVNLEPASEIKMDREQLRLIAWELIANRGGSRPPAAIPLASPGVVDA
Mflv_2585|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5509|M.smegmatis_MC2_155 -----------------MSLAATSSGICVSFGTNLRKDQQMTCEHAPLTT
Mb0930|M.bovis_AF2122/97 DASRLAVSWFGHSTALLEIDGYRVLTDPVWSDRCSPSDVVG---PQRLHP
Rv0906|M.tuberculosis_H37Rv DASRLAVSWFGHSTALLEIDGYRVLTDPVWSDRCSPSDVVG---PQRLHP
MAV_1029|M.avium_104 DPSRLAVSWFGHSTALLEIDGYRVLTDPVWSERCSPSDIVG---PQRLHP
Mflv_2585|M.gilvum_PYR-GCK --MQIDVTFIGTATTLIRCGNITVLTDPNFLHRGQRAYLGYGLWSKRRRA
MSMEG_5509|M.smegmatis_MC2_155 RRLPFGVAVLGGPTTVVDIGGHRLICDPTFDP---PTDYGY---LQKLAG
: *: :* .*::: .. :: ** : : ::
Mb0930|M.bovis_AF2122/97 PPVQLAALPAVDAVVISHDHYDHLDIDTVVALVGMQRAPFLVPLGVGAHL
Rv0906|M.tuberculosis_H37Rv PPVQLAALPAVDAVVISHDHYDHLDIDTVVALVGMQRAPFLVPLGVGAHL
MAV_1029|M.avium_104 PPIQLEGLPAVDAVVISHDHYDHLDIDTVRALARTQRAPFFVPLGVGAHL
Mflv_2585|M.gilvum_PYR-GCK PGLTPEELPPIDAIVLSHMHGDHWDRVAERSLDRELPVITTHEAAKALHR
MSMEG_5509|M.smegmatis_MC2_155 PAADESAVGDVDVALVSHDLHPDNLDVAGRAFALSAPVLLTTATAATRLG
* : :*. ::** . : :: . .
Mb0930|M.bovis_AF2122/97 RSWGVPQDRIVELDWNQSAQVDELTVVCVPARHFSG---RFLSRNTTLWA
Rv0906|M.tuberculosis_H37Rv RSWGVPQDRIVELDWNQSAQVDELTVVCVPARHFSG---RFLSRNTTLWA
MAV_1029|M.avium_104 RRWGIPEQRIVELDWHQSGKVDELTVTCVPARHFSG---RFLDRNTTLWA
Mflv_2585|M.gilvum_PYR-GCK KGFTEAVGLSTWDPFVLTKDGETLTVTAMPGRHAPTPVSRLLPPVMGSML
MSMEG_5509|M.smegmatis_MC2_155 ------APSVPLAPWTTWESPDGLAITAVPARHGPADGELNEEGFVNCEV
: . : *::..:*.** .
Mb0930|M.bovis_AF2122/97 SWAFVG--PNHRAYFGGDTGYTKSFTQIGADHGPFDLTLLPIG----AYN
Rv0906|M.tuberculosis_H37Rv SWAFVG--PNHRAYFGGDTGYTKSFTQIGADHGPFDLTLLPIG----AYN
MAV_1029|M.avium_104 SWALAG--PTHRAYFGGDTGYTKTFAQTGAEHGPFDLTLLPIG----AYN
Mflv_2585|M.gilvum_PYR-GCK DFDSGGGSSRRRVYISGDTLLIDELDEIPVRFNAIDAGVLHLGGTRLPFG
MSMEG_5509|M.smegmatis_MC2_155 IGFVVTAPDSPTVYLSGDNASVEIVREIRERIGGIDCAVLFAG---SASV
.*:.**. . . : . :* :* * .
Mb0930|M.bovis_AF2122/97 TAWPDIHMNPEEAVRAHLDVTDSGSGMLVPVHWGTFRLAPHPWGEPVERL
Rv0906|M.tuberculosis_H37Rv TAWPDIHMNPEEAVRAHLDVTDSGSGMLVPVHWGTFRLAPHPWGEPVERL
MAV_1029|M.avium_104 TAWPDVHMNPEEAVRAHRDVTNDG--LLVPIHWGTFRLAPHPWAEPVERL
Mflv_2585|M.gilvum_PYR-GCK DRLPVGLTVTMDGRQGAETVERLHLPRVIPVHFDDYRVFASPLADFHREM
MSMEG_5509|M.smegmatis_MC2_155 PAKFDGRPLSLTAERAAAAAEVLGSPHVVVAHQDGWAHFRQGAADTAAAF
. . :. . :: * . : .: :
Mb0930|M.bovis_AF2122/97 LAAAEPEHVTVAVPLPGQRVDPTGPMRLHPWWRL
Rv0906|M.tuberculosis_H37Rv LAAAEPEHVTVAVPLPGQRVDPTGPMRLHPWWRL
MAV_1029|M.avium_104 LAAAEPAKVKVAVPVPGQRIDPAAPQKPDPWWRL
Mflv_2585|M.gilvum_PYR-GCK SRRGLADRVVTVE--RGQTVSL------------
MSMEG_5509|M.smegmatis_MC2_155 HAAGIGDRLCSAP--LGHMCAI------------
. :: . *: