For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVRRALRLAAGTASLAAGTWLLRALHGTPAALGADAASIRAVSEQSPNYRDGAFVNLDPASMFTLDREEL RLIVWELVARHSASRPAAPIPLASPNIYRGDASRLAVSWFGHSTALLEIDGYRVLTDPVWSDRCSPSDVV GPQRLHPPPVQLAALPAVDAVVISHDHYDHLDIDTVVALVGMQRAPFLVPLGVGAHLRSWGVPQDRIVEL DWNQSAQVDELTVVCVPARHFSGRFLSRNTTLWASWAFVGPNHRAYFGGDTGYTKSFTQIGADHGPFDLT LLPIGAYNTAWPDIHMNPEEAVRAHLDVTDSGSGMLVPVHWGTFRLAPHPWGEPVERLLAAAEPEHVTVA VPLPGQRVDPTGPMRLHPWWRL
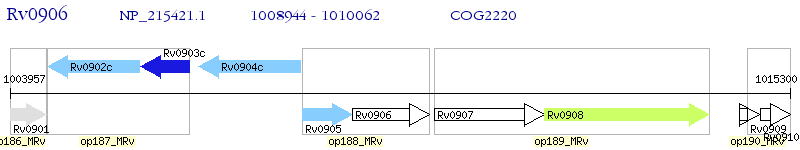
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0906 | - | - | 100% (372) | hypothetical protein Rv0906 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0930 | - | 0.0 | 100.00% (372) | hypothetical protein Mb0930 |
| M. gilvum PYR-GCK | Mflv_1756 | - | 1e-142 | 62.90% (372) | beta-lactamase domain-containing protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0960 | - | 1e-131 | 59.62% (369) | hypothetical protein MAB_0960 |
| M. marinum M | MMAR_4624 | - | 0.0 | 82.80% (372) | hypothetical protein MMAR_4624 |
| M. avium 104 | MAV_1029 | - | 1e-178 | 78.17% (371) | metallo-beta-lactamase family protein |
| M. smegmatis MC2 155 | MSMEG_5638 | - | 1e-148 | 66.58% (374) | metallo-beta-lactamase family protein |
| M. thermoresistible (build 8) | TH_0095 | - | 1e-154 | 67.66% (368) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0235 | - | 0.0 | 81.72% (372) | hypothetical protein MUL_0235 |
| M. vanbaalenii PYR-1 | Mvan_4991 | - | 1e-146 | 63.51% (370) | hypothetical protein Mvan_4991 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0906|M.tuberculosis_H37Rv -MVRRALRLAAGTAS---LAAGTWLLRALHGTPAALGADAASIRAVSEQS
Mb0930|M.bovis_AF2122/97 -MVRRALRLAAGTAS---LAAGTWLLRALHGTPAALGADAASIRAVSEQS
MMAR_4624|M.marinum_M -MVGRALRLAAGTAS---LAAGGWLMRALHGAPAALGADPAAIEAASDGS
MUL_0235|M.ulcerans_Agy99 -MVGRALRLAAGTAS---LAAGGWLMRALRGAPAALGADPAAIEAASDGS
MAV_1029|M.avium_104 --MRGTLRLVAGTAS---LAAGGWLVRALHGAPDALGADPASIAAVTSGS
MSMEG_5638|M.smegmatis_MC2_155 -MLRAALRLTTRSAG---LLAGGWLLGALRGTYASVGAQPRDIKPVATRS
TH_0095|M.thermoresistible__bu -MSRSWLGLMRQAAGVATVLGGGWLIRALRGAPAAVGADRRDIRPVAQRS
Mflv_1756|M.gilvum_PYR-GCK MVVRTALRFGFGTTS---LLAGGWVLRALHGTPASLGATPAEIAPVARRS
Mvan_4991|M.vanbaalenii_PYR-1 MEVRTALRFGFGTAS---LFAGGWVLRALQGTPAALGATPPEIAPVARRS
MAB_0960|M.abscessus_ATCC_1997 ---MRALWVGAGAAI-----ASTWIGRALRDVPQTLGASKGRIAEVAKGS
* . :: .. *: **:.. ::** * .: *
Rv0906|M.tuberculosis_H37Rv PNYRDGAFVNLDPAS-MFTLDREELRLIVWELVARHSASRPAAPIPLASP
Mb0930|M.bovis_AF2122/97 PNYRDGAFVNLDPAS-MFTLDREELRLIVWELVARHSASRPAAPIPLASP
MMAR_4624|M.marinum_M PNYRDGVFVNLDPAS-VFTLNREEMRLLAWELLANRGGSRPAKPIPLAAP
MUL_0235|M.ulcerans_Agy99 PYYRDGVFVNLDPAS-VFTLNREEMRLLAWELLANRGGSRPAKPIPLAAP
MAV_1029|M.avium_104 PHFRDGAFVNLEPAS-EIKMDREQLRLIAWELIANRGGSRPPAAIPLASP
MSMEG_5638|M.smegmatis_MC2_155 PQFDGKVFVNIEPASPGINLDREERRRLFAEMFGSKGASWPPGEIPLAEP
TH_0095|M.thermoresistible__bu PNYRDGAFVNLDPAS-MISLDRQQQWVLAREFLDGRAATRPAAPIPLVTP
Mflv_1756|M.gilvum_PYR-GCK PHYKDGRFVNLESSSSGLPMDRELQRKLLRDLANAGSAGKPPRPIPLKEP
Mvan_4991|M.vanbaalenii_PYR-1 PHYQDGKFVNLEPPS-GITMDRDLQRMLLRDLANFASQGTPPGPIPLADP
MAB_0960|M.abscessus_ATCC_1997 PQYRGGTFHNAEPAR-QFSPDAETT-TVVWDVLTRRGVGTPKGQVPLAVP
* : . * * :.. : : : : :. . * :** *
Rv0906|M.tuberculosis_H37Rv NIYRG--DASRLAVSWFGHSTALLEIDGYRVLTDPVWSDRCSPSDVVGPQ
Mb0930|M.bovis_AF2122/97 NIYRG--DASRLAVSWFGHSTALLEIDGYRVLTDPVWSDRCSPSDVVGPQ
MMAR_4624|M.marinum_M QVYQG--DASRLAVSWFGHSTAMVEIDGYRVLTDPVWSDRCSPSDLVGPQ
MUL_0235|M.ulcerans_Agy99 QVYQG--DASRLAVSWFGHSTAMVEIDGYRVLTDPVWSDRCSPSDLVGPQ
MAV_1029|M.avium_104 GVVDA--DPSRLAVSWFGHSTALLEIDGYRVLTDPVWSERCSPSDIVGPQ
MSMEG_5638|M.smegmatis_MC2_155 TEAET-AEAKPCAVSWYGHSSALIEVDGYRVLTDPIWSDRCSPSRVVGPR
TH_0095|M.thermoresistible__bu NPADHGVGAADLAVRWYGHSTAMVEVDGYRVLTDPIWSRRCSPSALVGPR
Mflv_1756|M.gilvum_PYR-GCK PTVEP--TGADAAASWYGHSSALIEVDGYRVLVDPVWSERCSPSRTVGPQ
Mvan_4991|M.vanbaalenii_PYR-1 PVVDP--TPAPAAASWYGHSSTLIEVDGYRVLADPVWSNRCSPSRAVGPQ
MAB_0960|M.abscessus_ATCC_1997 ELSGP---PADLAATWFGHSSVLVEIDGYRVLTDPVWSDRCSPSRVVGPH
*. *:***:.::*:******.**:** ***** ***:
Rv0906|M.tuberculosis_H37Rv RLHPPPVQLAALPAVDAVVISHDHYDHLDIDTVVALVGMQRAPFLVPLGV
Mb0930|M.bovis_AF2122/97 RLHPPPVQLAALPAVDAVVISHDHYDHLDIDTVVALVGMQRAPFLVPLGV
MMAR_4624|M.marinum_M RLHPPPVQLDGLPAVDAVVISHDHYDHLDIDTVIALVRSQRAPFFVPLGV
MUL_0235|M.ulcerans_Agy99 RLHPPPVQLDGLPAVDAVVISHDHYDHLDIDTVIALVRSQRAPFFVPLGV
MAV_1029|M.avium_104 RLHPPPIQLEGLPAVDAVVISHDHYDHLDIDTVRALARTQRAPFFVPLGV
MSMEG_5638|M.smegmatis_MC2_155 RLHAPPVPLEALPAVDAVVISHDHYDHLDIDTVRGLARTQRAPFLVPLGI
TH_0095|M.thermoresistible__bu RLHEPPTPLEALPAVDAVLISHDHYDHLDMDTVVRLARTQRAPFLVPLGV
Mflv_1756|M.gilvum_PYR-GCK RMHDVPLLLESLPAVDAVVISHDHYDHLDIDTIIALARTQRAPFVVPLGI
Mvan_4991|M.vanbaalenii_PYR-1 RMHDVPVLIEALPAVDAVIISHDHYDHLDIDTIVALAHTQRAPFVVPLGI
MAB_0960|M.abscessus_ATCC_1997 RQHPVPVELSALPALDAVVISHDHYDHLDMDSIIALTRSQNAVFVVPLGV
* * * : .***:***:**********:*:: *. *.* *.****:
Rv0906|M.tuberculosis_H37Rv GAHLRSWGVPQDRIVELDWNQSAQVDELTVVCVPARHFSGRFLSRNTTLW
Mb0930|M.bovis_AF2122/97 GAHLRSWGVPQDRIVELDWNQSAQVDELTVVCVPARHFSGRFLSRNTTLW
MMAR_4624|M.marinum_M GAHLRSWGVPEDRIIELDWHQSAQVGELSVICMPARHFSGRFMSRNNTLW
MUL_0235|M.ulcerans_Agy99 GAHLRSWGVPEDRIIEFDWHQSAQVGELSVICMPARHFSGRFMSRNNTLW
MAV_1029|M.avium_104 GAHLRRWGIPEQRIVELDWHQSGKVDELTVTCVPARHFSGRFLDRNTTLW
MSMEG_5638|M.smegmatis_MC2_155 GAHLRKWGIPEARIVELDWNESHRIGDLTIVCAPARHFSGRFLQRNTTLW
TH_0095|M.thermoresistible__bu GAHLRTWRIPEHRIVELDWYESRRIGDLRITCTPARHFSGRLFARNTTLW
Mflv_1756|M.gilvum_PYR-GCK GAHLRKWGIPSRRIVELDWHEGHRIGDLTLICTPARHFSGRLFSRDTTLW
Mvan_4991|M.vanbaalenii_PYR-1 GAHLRKWGIPESRIVELDWNEAHRIGDLTLICTPARHFSGRLFARDSTLW
MAB_0960|M.abscessus_ATCC_1997 GAHLRGWGVSPARVIELDWDQSHQLGKLTLTCTQARHFSGRSITRNTTLW
***** * :. *::*:** :. ::..* : * ******* : *:.***
Rv0906|M.tuberculosis_H37Rv ASWAFVGPNHRAYFGGDTGYTKSFTQIGADHGPFDLTLLPIGAYNTAWPD
Mb0930|M.bovis_AF2122/97 ASWAFVGPNHRAYFGGDTGYTKSFTQIGADHGPFDLTLLPIGAYNTAWPD
MMAR_4624|M.marinum_M ASWAFVGPTHRAYFGGDTGYTKSFAEIGADHGPFDLTLMPIGAYNTGWPD
MUL_0235|M.ulcerans_Agy99 ASWAFVGPTHRAYFGGDTGYTKSFAEIGADHGPFDLTLMPIGAYSTGWPD
MAV_1029|M.avium_104 ASWALAGPTHRAYFGGDTGYTKTFAQTGAEHGPFDLTLLPIGAYNTAWPD
MSMEG_5638|M.smegmatis_MC2_155 ASWVVIGPGHRAFFGGDTGYTKSFSEIGSEHGPFDLTLLPVGAYNPGWPD
TH_0095|M.thermoresistible__bu ASWVIAGPRHRAFFGGDTGYTKSFTEIGVDHGPFDLTLLPIGAYHPAWPD
Mflv_1756|M.gilvum_PYR-GCK ASWVITGPSHKAFFGGDTGYTKSFADIGAQYGPFDLTLMPIGAYHPAFAD
Mvan_4991|M.vanbaalenii_PYR-1 SSWVVTGPTHKAFFGGDTGYTKSFAEIGAEHGPFDLTLLPIGAYHPAFAD
MAB_0960|M.abscessus_ATCC_1997 ASWAFAGPKHKVFFGGDTGYTKAFKVIGDTYGPFDLTLLPVGAYNTSWAD
:**.. ** *:.:*********:* * :*******:*:*** ..:.*
Rv0906|M.tuberculosis_H37Rv IHMNPEEAVRAHLDVTDSGSGMLVPVHWGTFRLAPHPWGEPVERLLAAAE
Mb0930|M.bovis_AF2122/97 IHMNPEEAVRAHLDVTDSGSGMLVPVHWGTFRLAPHPWGEPVERLLAAAE
MMAR_4624|M.marinum_M IHMNPEEAVRAHLDVSDTGSGLLVPIHWGTFRLAPHPWAEPVERLLKAAE
MUL_0235|M.ulcerans_Agy99 IHMNPEEAVRAHLDVSDTGSGLLVPIHWGTFRLAPHPWAEPVERLLKAAE
MAV_1029|M.avium_104 VHMNPEEAVRAHRDVTN--DGLLVPIHWGTFRLAPHPWAEPVERLLAAAE
MSMEG_5638|M.smegmatis_MC2_155 IHMNPEESVRAHLDMTES--GLLVPIHWATFRLAPHPWAEPVRRLLAAAD
TH_0095|M.thermoresistible__bu IHMNPEEAVRAHLDMAEADSGLLVPIHWATFRLAPHPWSEPVERLLAAAD
Mflv_1756|M.gilvum_PYR-GCK IHMNPEEAVRAHLDLADPDASVLVPVHWATFRLAPHPWAEPVERLVTAAE
Mvan_4991|M.vanbaalenii_PYR-1 IHMNPEEAVRAHLDLADVDNSLMVPIHWATFRLAPHPWAEPAERLLTAAD
MAB_0960|M.abscessus_ATCC_1997 IHMNPEEAVQTHLDLATA-QAPLLPIHWATFNLALHPWAEPIERVLAAAE
:******:*::* *:: . ::*:**.**.** ***.** .*:: **:
Rv0906|M.tuberculosis_H37Rv PEHVTVAVPLPGQRVDPTGPMRLHPWWRL----
Mb0930|M.bovis_AF2122/97 PEHVTVAVPLPGQRVDPTGPMRLHPWWRL----
MMAR_4624|M.marinum_M SDQVQVAVPLPGQRIDPAGPLRFNPWWRL----
MUL_0235|M.ulcerans_Agy99 SDQVQVAVPLPGQGVDPAGPLRFNPWWRL----
MAV_1029|M.avium_104 PAKVKVAVPVPGQRIDPAAPQKPDPWWRL----
MSMEG_5638|M.smegmatis_MC2_155 PAGVQVAVPKPGQRVDGGSP-VLDPWWQL----
TH_0095|M.thermoresistible__bu PARVPVAVPRPGERVDRDSAGASDQWWRPADGR
Mflv_1756|M.gilvum_PYR-GCK LDGVRLAVPIPGERVLPGST--FEPWWRP----
Mvan_4991|M.vanbaalenii_PYR-1 AERVRVAVPMPGQRVVPEST--FDPWWRF----
MAB_0960|M.abscessus_ATCC_1997 EQGVTVVAPRPGQRADARQPAAPDGWWRLT---
* :..* **: . . **: