For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAPQWHLGPVTVEVHGLFVALAVFAAVLVFSHEARRRGAVNEQSIVAVAGALIGGAIGMRLSGWAQHLDL SANPSLAQAWEFGSRSILGGLLGAYIGVHVAKRLGGYRGKTGDLFAPAVALGMAIGRIGCLLTEAPGRPT NLPWGIHAPASTPECPACLTGQAMHPSFVYEIVFQLVAFAVLLSLRNRITHPGELFVIYIAAYAVFRFFV EFFRANETVWLDLTRPQWFLLPTLLIIGFRLCYGYRHGYYRRAARSEEVPV
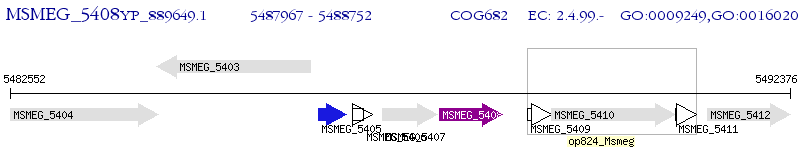
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5408 | lgt | - | 100% (261) | prolipoprotein diacylglyceryl transferase |
| M. smegmatis MC2 155 | MSMEG_3222 | - | 2e-23 | 30.74% (231) | prolipoprotein diacylglyceryl transferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1640 | lgt | 1e-17 | 31.33% (233) | prolipoprotein diacylglyceryl transferase |
| M. gilvum PYR-GCK | Mflv_3597 | - | 2e-21 | 30.08% (236) | prolipoprotein diacylglyceryl transferase |
| M. tuberculosis H37Rv | Rv1614 | lgt | 1e-17 | 31.33% (233) | prolipoprotein diacylglyceryl transferase |
| M. leprae Br4923 | MLBr_01274 | - | 2e-18 | 30.04% (233) | putative prolipoprotein diacylglyceryl transferase |
| M. abscessus ATCC 19977 | MAB_2642c | - | 1e-21 | 28.63% (255) | prolipoprotein diacylglyceryl transferase |
| M. marinum M | MMAR_2416 | lgt | 2e-19 | 30.77% (234) | prolipoprotein diacylglyceryl transferases Lgt |
| M. avium 104 | MAV_3172 | - | 8e-21 | 31.33% (233) | prolipoprotein diacylglyceryl transferase |
| M. thermoresistible (build 8) | TH_1073 | lgt | 2e-22 | 30.30% (231) | Possible prolipoprotein diacylglyceryl transferases Lgt |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2819 | - | 5e-25 | 32.81% (253) | prolipoprotein diacylglyceryl transferase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2642c|M.abscessus_ATCC_199 MTVVNLAYIPSPPQGVWHLGPIPIRAYALCIIAGIVAALIIGDRRWEQRG
Mvan_2819|M.vanbaalenii_PYR-1 MTTTVLAYIPSPSQGVWHLGPFPVRAYALFIIVGIIAALVIGDRRWVARG
Mflv_3597|M.gilvum_PYR-GCK MTTTVLAYIPSPPQGVWYLGPFPLRAYALCIIVGIVAALVIGDRRWVARG
Mb1640|M.bovis_AF2122/97 -MRMLPSYIPSPPRGVWYLGPLPVRAYAVCVITGIIVALLIGDRRLTARG
Rv1614|M.tuberculosis_H37Rv -MRMLPSYIPSPPRGVWYLGPLPVRAYAVCVITGIIVALLIGDRRLTARG
MLBr_01274|M.leprae_Br4923 MTRMLPGYFPSPPRGVWHLGPLPIRAYALLIILGIVAALVVGGRCWEARG
MMAR_2416|M.marinum_M MTATVLAYFPSPPRGVWHLGPLPIRAYAFFIIIGIVMGLMIGDRRFAARG
MAV_3172|M.avium_104 -MTKLLAYFPSPPQGVWHLGPVPIRAYALFIIAGIVAALLIGDRRWEARG
TH_1073|M.thermoresistible__bu -VTTTLAYIPSPAQGVWQLGPIPIRAYALCIIAGIVVALVLGDRRWAARG
MSMEG_5408|M.smegmatis_MC2_155 ------------MAPQWHLGPVTVEVHGLFVALAVFAAVLV----FSHEA
* ***..:..:.. : .:. .::: ..
MAB_2642c|M.abscessus_ATCC_199 GERGVIYDIALWAVPFGLVGGRLYHVMTDWQTYFGAHGKGIGKAIAVWEG
Mvan_2819|M.vanbaalenii_PYR-1 GESGVIYDIALWAVPFGLIGGRLYHVITDWPTYFGEGGAGPAAALRIWDG
Mflv_3597|M.gilvum_PYR-GCK GEPGVIYDIALWAVPFGLIGGRLYHVITDWPTYFGDGGAGPAAALRIWDG
Mb1640|M.bovis_AF2122/97 GERGMTYDIALWAVPFGLIGGRLYHLATDWRTYFGDGGAGLAAALRIWDG
Rv1614|M.tuberculosis_H37Rv GERGMTYDIALWAVPFGLIGGRLYHLATDWRTYFGDGGAGLAAALRIWDG
MLBr_01274|M.leprae_Br4923 GERDVTYDIALWAVPFGLVGGRLYHLATDWRTYFGQNGAGLGAALRIWDG
MMAR_2416|M.marinum_M GERGVIYDIALWMVPFGLIGGRLYHLSTDWQTYFGEGGAGLGAALRIWDG
MAV_3172|M.avium_104 GERGVIYDIALWTVPFGLVGGRLYHLATDWRTYWGPGGAGFGAAVRIWDG
TH_1073|M.thermoresistible__bu GERGVIYDIALWAVPFGLIGGRVYHVLTDWRTYFGPDGAGFVAALRIWDG
MSMEG_5408|M.smegmatis_MC2_155 RRRGAVNEQSIVAVAGALIGGAIGMRLSGWAQHLDLSAN--PSLAQAWEF
. . : :: *. .*:** : :.* : . . *:
MAB_2642c|M.abscessus_ATCC_199 G-LGIWGAVALGGVGAWIACRRRGIPLPAFADAIAPGIVLAQAIGRLGNY
Mvan_2819|M.vanbaalenii_PYR-1 G-LGIWGAVALGGIGAWIACRRRGIPLPAFGDAIAPGIVLAQAIGRLGNY
Mflv_3597|M.gilvum_PYR-GCK G-LGIWGAVALGAVGAWIACRRRGIPLPAFGDAIAPGIVLAQAIGRLGNY
Mb1640|M.bovis_AF2122/97 G-LGIWGAVTLGVMGAWIGCRRCGIPLPVLLDAVAPGVVLAQAIGRLGNY
Rv1614|M.tuberculosis_H37Rv G-LGIWGAVTLGVMGAWIGCRRCGIPLPVLLDAVAPGVVLAQAIGRLGNY
MLBr_01274|M.leprae_Br4923 G-LGIWGAVALGCVGAWLGCRRHRIPLPAFGDALAPGIILAQAIGRLGNY
MMAR_2416|M.marinum_M G-LGIWGAVTLGTVGAWIGCRRRGIPLPAFLDAVAPGVVMGQAIGRLGNY
MAV_3172|M.avium_104 G-LGIWGAVALGAVGAWIGCRRHGIPLPAFADALAPGIILAQAIGRLGNY
TH_1073|M.thermoresistible__bu G-LGIWGAVALGGVGAWIACRRHGIPLPAFGDAIAPGIVLAQAIGRLGNY
MSMEG_5408|M.smegmatis_MC2_155 GSRSILGGLLGAYIGVHVAKRLGGYRG-KTGDLFAPAVALGMAIGRIG-C
* .* *.: . :*. :. * * .**.: :. ****:*
MAB_2642c|M.abscessus_ATCC_199 FNQELTGGPTTLPWGLELFYRRSADGTVDVLNLNGVSTGEVAQVVHPTFL
Mvan_2819|M.vanbaalenii_PYR-1 FNQELFGRETTVPWGLEIYERVNAAGVPDSLN--GVSTGELIQVVHPTFL
Mflv_3597|M.gilvum_PYR-GCK FNQELYGRETTVPWGLEIYERRNSAGVLDSLN--GISTGELIGVVHPTFL
Mb1640|M.bovis_AF2122/97 FNQELYGRETTMPWGLEIFYRRDPSGFDVPNSLDGVSTGQVAFVVQPTFL
Rv1614|M.tuberculosis_H37Rv FNQELYGRETTMPWGLEIFYRRDPSGFDVPNSLDGVSTGQVAFVVQPTFL
MLBr_01274|M.leprae_Br4923 FNQELYGRETTMPWGLEVFYRRDPAGYMDPHSLDGVSTGQLAFVVQPTFL
MMAR_2416|M.marinum_M FNQELYGRETTVPWGLEIFYRRDPSGFVDVHSLDGVSTGQVAVVVQPTFL
MAV_3172|M.avium_104 FNQELYGRETTLPWGLEIFYRRDPSGYIDPHSLDGVSTGQVALVVQPTFL
TH_1073|M.thermoresistible__bu FNQELYGRPTTLPWGLEIYERRNASGALDPLN--GTSTGELLAVVHPTFL
MSMEG_5408|M.smegmatis_MC2_155 LLTEAPGRPTNLPWGIHAPASTP-----------ECPACLTGQAMHPSFV
: * * *.:***:. .: .::*:*:
MAB_2642c|M.abscessus_ATCC_199 YELLWNVLVFVLLIVVDRRFKIGHGRLFAMYVAAYCVGRFCVELMRVDPA
Mvan_2819|M.vanbaalenii_PYR-1 YELLWNLLVFAVLILVDRRFKIGHGRLFALYVAGYCVGRFWIELMRSDTA
Mflv_3597|M.gilvum_PYR-GCK YELLWNLLVFAVLILVDRRFKIGHGRLFALYVAGYCAGRFWIELMRSDTA
Mb1640|M.bovis_AF2122/97 YELIWNVLVFVALIYIDRRFIIGHGRLFGFYVAFYCAGRFCVELLRDDPA
Rv1614|M.tuberculosis_H37Rv YELIWNVLVFVALIYIDRRFIIGHGRLFGFYVAFYCAGRFCVELLRDDPA
MLBr_01274|M.leprae_Br4923 YELIWNVLVFFALIYVDRWFTLGHGRLFATYVAAYCIGRFCVELLRDDAA
MMAR_2416|M.marinum_M YELIWNLLVFAALIYLDRRFTIGHGRLFASYVALYCVGRFCVELLRDDTA
MAV_3172|M.avium_104 YELLWNLLIFVALLYADRRLTLGHGRLFALYVAGYCVGRFCVELLRDDTA
TH_1073|M.thermoresistible__bu YEMLWNVLVFVVLIWADRRFRLGHGRLFALYVAGYCVGRFWVELLRSDFA
MSMEG_5408|M.smegmatis_MC2_155 YEIVFQLVAFAVLLSLRNRITH-PGELFVIYIAAYAVFRFFVEFFRANET
**:::::: * *: . : *.** *:* *. ** :*::* : :
MAB_2642c|M.abscessus_ATCC_199 TQFGGIRVNSFTSTLVFVGAVAYILLAPKGREDPATLGG-----------
Mvan_2819|M.vanbaalenii_PYR-1 TLLAGIRVNSFTSTFVFIGAVVYIMAAPKGREDPSSLKGNQDAEARVEDL
Mflv_3597|M.gilvum_PYR-GCK TLLAGIRVNTFTATFVFVAAVVYVIVAPKGREAPESLRGNQDAESDVEDL
Mb1640|M.bovis_AF2122/97 TLIAGIRINSFTSTFVFIGAVVYIILAPKGREAPGALRGSE---------
Rv1614|M.tuberculosis_H37Rv TLIAGIRINSFTSTFVFIGAVVYIILAPKGREAPGALRGSE---------
MLBr_01274|M.leprae_Br4923 THIAGIRINSFTSTFVFIGAVVYIILAPKGREEPENLCRAE---------
MMAR_2416|M.marinum_M THIAGIRINSFTSTFVFIGAVVYIILAPKGREDPASVRGTA---------
MAV_3172|M.avium_104 THIAGIRINSFTSTFVFIGAVVYLMAAPKGREDPESLRGNQ---------
TH_1073|M.thermoresistible__bu THIAGIRINSFTASFVFIGAVVYIMVAPKGREDPATLGGPV---------
MSMEG_5408|M.smegmatis_MC2_155 VWLDLTRPQWFLLPTLLIIGFRLCYGYRHGYYRRAARSEEVPV-------
. : * : * . ::: .. :*
MAB_2642c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_2819|M.vanbaalenii_PYR-1 AKDVAVGSATGIVAAATVADEDEAMEPADDDDAAEVEALVAELEASEPET
Mflv_3597|M.gilvum_PYR-GCK AKDVALASAVGVVAAATVAGEDE------------------KRDGGGSPP
Mb1640|M.bovis_AF2122/97 --------------------------------------------------
Rv1614|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01274|M.leprae_Br4923 --------------------------------------------------
MMAR_2416|M.marinum_M --------------------------------------------------
MAV_3172|M.avium_104 --------------------------------------------------
TH_1073|M.thermoresistible__bu --------------------------------------------------
MSMEG_5408|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2642c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_2819|M.vanbaalenii_PYR-1 EGLGSVEGDEVAEAVEVAPEVHEAAVEGEAEAEELASDVEGAPEPEGVEA
Mflv_3597|M.gilvum_PYR-GCK GGLGSVEGDEALEAAG---------------------------------G
Mb1640|M.bovis_AF2122/97 --------------------------------------------------
Rv1614|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01274|M.leprae_Br4923 --------------------------------------------------
MMAR_2416|M.marinum_M --------------------------------------------------
MAV_3172|M.avium_104 --------------------------------------------------
TH_1073|M.thermoresistible__bu --------------------------------------------------
MSMEG_5408|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2642c|M.abscessus_ATCC_199 ----TPSSADG-------------------------GDDTAET-------
Mvan_2819|M.vanbaalenii_PYR-1 AEAEAESPVDGAEAESEVVEGDVESVTESEGLGSVEGDEVAEAVEVAPEV
Mflv_3597|M.gilvum_PYR-GCK ASAANESAVEG-------------------------AADAADLVALLEAS
Mb1640|M.bovis_AF2122/97 --YVVDEALER---------------------------EPAELAAAAVAS
Rv1614|M.tuberculosis_H37Rv --YVVDEALER---------------------------EPAELAAAAVAS
MLBr_01274|M.leprae_Br4923 --YVAREIPEP---------------------------ESATERATVAS-
MMAR_2416|M.marinum_M --AAAEEVPEP---------------------------EPAGEVATAAAP
MAV_3172|M.avium_104 --YVEEEPAEP---------------------------EPATVAATTEA-
TH_1073|M.thermoresistible__bu ---QTDAGADT---------------------------GAAGADAEVDT-
MSMEG_5408|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2642c|M.abscessus_ATCC_199 ----------------EATADTEDTEDTEDGVTDAPEADATDKSVDESAD
Mvan_2819|M.vanbaalenii_PYR-1 HEVVVEGEAEAEALAADAEAEAEAEAEAEDLAADVEAESPVDGAEDESEA
Mflv_3597|M.gilvum_PYR-GCK EDVEVADGAVEPQEVDAPPVPDEAGGLGAVEGEEAAEAAAGAGAARASVS
Mb1640|M.bovis_AF2122/97 AASAVG--PVGPGEPNQPDDVAEAVKAEVAEVTDEVAAESVVQVADRDGE
Rv1614|M.tuberculosis_H37Rv AASAVG--PVGPGEPNQPDDVAEAVKAEVAEVTDEVAAESVVQVADRDGE
MLBr_01274|M.leprae_Br4923 ---------------------TYATTTAVPVSADEEFAETN---------
MMAR_2416|M.marinum_M AGDTEGDEPTGLAETAKAAEVAEPETVGSAVAGDESTADTEASDAVESEV
MAV_3172|M.avium_104 ---------------------ATEGVAAPADGAEAAGADATAQRPEESAE
TH_1073|M.thermoresistible__bu -------------EVDTEKEDVAAAVRGVAGAGAXXXRHIARRRGRRRGE
MSMEG_5408|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2642c|M.abscessus_ATCC_199 NSGIVEK-------------------------------------------
Mvan_2819|M.vanbaalenii_PYR-1 ESEVVEGDVESVTESEGLGS-------VEGDEVAEAVEVAPEVHEAAVEG
Mflv_3597|M.gilvum_PYR-GCK DDAEASNDADAETEAAGLGS-------VDGDEVAAAVEDAPEVHEAAVEG
Mb1640|M.bovis_AF2122/97 STPAVEETSEADIEREQPGD-----------------LAGQAPAAHQVDA
Rv1614|M.tuberculosis_H37Rv STPAVEETSEADIEREQPGD-----------------LAGQAPAAHQVDA
MLBr_01274|M.leprae_Br4923 --------------------------------------------------
MMAR_2416|M.marinum_M GEAAEAEAPEAEAEEAEVGEVEVAEAVEGEVEEAEEAEPGEVEVAEAVEG
MAV_3172|M.avium_104 PDVEKPESEETEAEAAEEPE-------------------SEETEAAEEPG
TH_1073|M.thermoresistible__bu RIRRRRRRQLVDRRRVQWLR-----------------RGIVHHIARGRAG
MSMEG_5408|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2642c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_2819|M.vanbaalenii_PYR-1 EAEAEELASDVEGAPEPEGVEAAEAEAESPVDG-----------AEAEAE
Mflv_3597|M.gilvum_PYR-GCK EAEAEELAAEAEVEAVAAEVAEVDADALTGADDDVDAETEAAVEGEAEAE
Mb1640|M.bovis_AF2122/97 EAASAAPEEPAALASEAHDETEP---------------------------
Rv1614|M.tuberculosis_H37Rv EAASAAPEEPAALASEAHDETEP---------------------------
MLBr_01274|M.leprae_Br4923 --------------------------------------------------
MMAR_2416|M.marinum_M EVEAAAEAEEAEEAEEAEEAEEAEEAEPGEIEVAEAVEGEVEAAAEAEEA
MAV_3172|M.avium_104 EPEAEEPEEPEAEEPEEPETEEP---------------------------
TH_1073|M.thermoresistible__bu QHLAAGRPRPLALAGQHPLGDRDLLLLGGQVGVR----------------
MSMEG_5408|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2642c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_2819|M.vanbaalenii_PYR-1 SEVVEGDVESVTESEGLGSVEGDEVAEAVEVAPEVHEVVVEGEAEAEALA
Mflv_3597|M.gilvum_PYR-GCK ELAAEADAEAQVAAEVA-EVDADALTGADDDVDAETEAEVEGEAEAEELA
Mb1640|M.bovis_AF2122/97 ---------EVPEKAAPIP-------------------------------
Rv1614|M.tuberculosis_H37Rv ---------EVPEKAAPIP-------------------------------
MLBr_01274|M.leprae_Br4923 --------------------------------------------------
MMAR_2416|M.marinum_M EVGEVEAVDEAAEAEAPEVGEIEAAEAVEGEVEVAAEAEEAEAGEDEPKE
MAV_3172|M.avium_104 ---------EADSDEEPEE-------------------------------
TH_1073|M.thermoresistible__bu --------RELAAAVGPPGAG-----------------------------
MSMEG_5408|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2642c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_2819|M.vanbaalenii_PYR-1 ADADADADAEAEDLAA----------DVEAESPVDGAEDESEAEPEAVEG
Mflv_3597|M.gilvum_PYR-GCK AEADAEAQVAAEVAEVDAETLTGADKDVDAEALTDADAETEAAGLGSVDG
Mb1640|M.bovis_AF2122/97 --------------------------------------------------
Rv1614|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01274|M.leprae_Br4923 --------------------------------------------------
MMAR_2416|M.marinum_M AEEVAEAEGSEAEDKTVQAIDGGQTGVDDAGEADAEPDDDSTAAEPEVVV
MAV_3172|M.avium_104 --------------------------------------------------
TH_1073|M.thermoresistible__bu --------------------------------------------------
MSMEG_5408|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2642c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_2819|M.vanbaalenii_PYR-1 D--VESVTESEGLGSVEGDEAAEAEELVGEVEADVEAETGEAAADEVAEV
Mflv_3597|M.gilvum_PYR-GCK DEVAAAVEDAPEVHEAAVEGESEAEELAAEADAEALAEVAEIDAEAVNDA
Mb1640|M.bovis_AF2122/97 --------DPAKPDELAVAG------------------------------
Rv1614|M.tuberculosis_H37Rv --------DPAKPDELAVAG------------------------------
MLBr_01274|M.leprae_Br4923 --------------------------------------------------
MMAR_2416|M.marinum_M EQADIEEIEPQEPEAEAVAAAEATDADEGDGEPAEPASDDLDAVAAEDAD
MAV_3172|M.avium_104 --------ESGEAPEQPVAE------------------------------
TH_1073|M.thermoresistible__bu --------SELQAPVIAVTG------------------------------
MSMEG_5408|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2642c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_2819|M.vanbaalenii_PYR-1 DAGAVTDTDDDAEGLGSVEGDEVAESAEDASEVHEAAVEREEAAEERAAE
Mflv_3597|M.gilvum_PYR-GCK DEDVDAEAEAEAAGLGSVDGDEVAAAVEDAPEVREAVEGVAEVDAEALTD
Mb1640|M.bovis_AF2122/97 -------------------PGDDPAEPDG---------------------
Rv1614|M.tuberculosis_H37Rv -------------------PGDDPAEPDG---------------------
MLBr_01274|M.leprae_Br4923 --------------------------------------------------
MMAR_2416|M.marinum_M VDVDATATEAEESGDESPEPAADSEEPEADESAPEDSAEDTSSGESDTQS
MAV_3172|M.avium_104 ------------------EPEPAPQQPET---------------------
TH_1073|M.thermoresistible__bu ------------------VDGPVSARLATGDLIPFGVGGRTG--------
MSMEG_5408|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2642c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_2819|M.vanbaalenii_PYR-1 AEADDA-SEPQPEEAADTEPAADGAVTRVESRSVATESQSGGLLRRMLRR
Mflv_3597|M.gilvum_PYR-GCK AEPEESDAEGEPEGPALPTSEHSPYTVVTESSSEPAQSGPAGLVRQWIRR
Mb1640|M.bovis_AF2122/97 -------IRRQDDFSS------------------RRRRWWRLRRRRQ---
Rv1614|M.tuberculosis_H37Rv -------IRRQDDFSS------------------RRRRWWRLRRRRQ---
MLBr_01274|M.leprae_Br4923 --------------------------------------------------
MMAR_2416|M.marinum_M ELEQSQTIEQGSDQTSAGDGEGPPPSAAVDGDTGPTRQGWRGRLRNRMRR
MAV_3172|M.avium_104 ------------------------------------KRRWGARLRDRLSG
TH_1073|M.thermoresistible__bu -------LTGHRQAAT------------------AEHSPGDSDFRDVYG-
MSMEG_5408|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2642c|M.abscessus_ATCC_199 ----
Mvan_2819|M.vanbaalenii_PYR-1 RRSR
Mflv_3597|M.gilvum_PYR-GCK RRNR
Mb1640|M.bovis_AF2122/97 ----
Rv1614|M.tuberculosis_H37Rv ----
MLBr_01274|M.leprae_Br4923 ----
MMAR_2416|M.marinum_M SGR-
MAV_3172|M.avium_104 R---
TH_1073|M.thermoresistible__bu ----
MSMEG_5408|M.smegmatis_MC2_155 ----