For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
THSTKLMSLLAPRAAAPADTRRTCSAGRRRLGVVELALPANALRDKAVARWVRDRSVAVEVRDGHELAIA QSAGVHPSRVTAHAEGLSIDELVFCSVGLRVGRVVVTRADHVDVLAVTRRRQAVLLGSLRRRLSRTVFDC ATVDVVGMYGDVDHAAHHSVSYPSAVGAMLAEMSRIRRDHGVMLTRIALGGRHLTFGAGAGELSEVSAAV DGTLDDACATLRFPRPSVVVSASPAV*
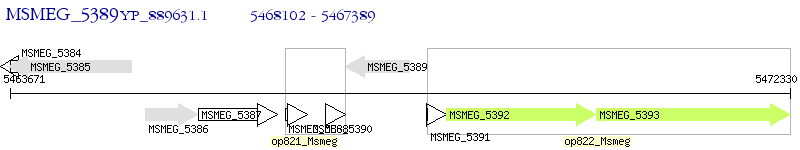
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5389 | - | - | 100% (237) | LysA protein |
| M. smegmatis MC2 155 | MSMEG_4958 | lysA | 4e-06 | 38.81% (67) | diaminopimelate decarboxylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1325 | lysA | 9e-07 | 35.00% (80) | diaminopimelate decarboxylase LysA |
| M. gilvum PYR-GCK | Mflv_2297 | - | 9e-06 | 24.46% (233) | diaminopimelate decarboxylase |
| M. tuberculosis H37Rv | Rv1293 | lysA | 9e-07 | 35.00% (80) | diaminopimelate decarboxylase LysA |
| M. leprae Br4923 | MLBr_01128 | lysA | 3e-05 | 31.25% (80) | diaminopimelate decarboxylase |
| M. abscessus ATCC 19977 | MAB_1156c | - | 3e-19 | 29.20% (250) | diaminopimelate decarboxylase LysA |
| M. marinum M | MMAR_4104 | lysA | 2e-06 | 25.90% (139) | diaminopimelate decarboxylase LysA |
| M. avium 104 | MAV_1160 | lysA | 4e-24 | 31.08% (251) | diaminopimelate decarboxylase |
| M. thermoresistible (build 8) | TH_2005 | lysA | 3e-06 | 28.78% (139) | PROBABLE DIAMINOPIMELATE DECARBOXYLASE LYSA (DAP |
| M. ulcerans Agy99 | MUL_3971 | lysA | 1e-06 | 25.90% (139) | diaminopimelate decarboxylase LysA |
| M. vanbaalenii PYR-1 | Mvan_2486 | - | 7e-45 | 42.46% (252) | Orn/DAP/Arg decarboxylase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4104|M.marinum_M MNVHPAGPRHAEESRHPESPQRPQSAEELLYLAPNVWPRNTTRDEQGCAR
MUL_3971|M.ulcerans_Agy99 MNVHPAGPRHAEESRHPESPQRPQSAEELLYLAPNVWPRNTARDEQGCAR
Mb1325|M.bovis_AF2122/97 -------------------------MNELLHLAPNVWPRNTTRDEVGVVC
Rv1293|M.tuberculosis_H37Rv -------------------------MNELLHLAPNVWPRNTTRDEVGVVC
MLBr_01128|M.leprae_Br4923 MNVHTAGPRHAEKTRHTATPQRVQPSDDLLRLASNVWPRNITRDETGVAC
Mflv_2297|M.gilvum_PYR-GCK MIAHPAGPRHAEEVPSAAAPERPQSAADVSRLAPNVWPQNTVRGDDGAVS
TH_2005|M.thermoresistible__bu VIAHPAGPRHAEEIHHGETPPRPQSPDELLLLAPNVWPRNTVRESDGVVS
MAB_1156c|M.abscessus_ATCC_199 MTLLDLLP----SLHNVARP----------RVDPAVWPVTTHLDEQGRMC
MAV_1160|M.avium_104 --MLDILP----SLGHAAPP----------RFDPAIWPVTAHPDEEGRLC
MSMEG_5389|M.smegmatis_MC2_155 ------------------------------------------------MT
Mvan_2486|M.vanbaalenii_PYR-1 ------------------------------------------------MT
MMAR_4104|M.marinum_M IAGVPLTELAREYG---TPLFVIDEDDFRSRCQEIAAAFGGGHNVNYAAK
MUL_3971|M.ulcerans_Agy99 IAGVPLTELAREYG---TPLFVIDEDDFRSRCQEIAAAFGGGHNVNYAAK
Mb1325|M.bovis_AF2122/97 IAGIPLTQLAQEYG---TPLFVIDEDDFRSRCRETAAAFGSGANVHYAAK
Rv1293|M.tuberculosis_H37Rv IAGIPLTQLAQEYG---TPLFVIDEDDFRSRCRETAAAFGSGANVHYAAK
MLBr_01128|M.leprae_Br4923 IAGNKLTDLAGEYG---TPLFVIDEDDFRFRCREIAAAFGGGENVHYAAK
Mflv_2297|M.gilvum_PYR-GCK IAGVPVADIAAEFG---TPVFVIDEADFRSRCRDISSAFGGGEYVHYAAK
TH_2005|M.thermoresistible__bu IAGVRVTDIADEYG---TPTFVIDEDDFRSRCREMAAAFGGGENVHYAAK
MAB_1156c|M.abscessus_ATCC_199 VGETALSEIADQFR---TPAYVLDEEDFRHRARRYRSTLR-EIEVAYAGK
MAV_1160|M.avium_104 VGGVPLGDVADEFG---TPVYVVDEADFRCRARRYRRALR-GVEVVYAGK
MSMEG_5389|M.smegmatis_MC2_155 HSTKLMSLLA-------PRAAAPADTR--RTCSAGRRRLG-VVELALPAN
Mvan_2486|M.vanbaalenii_PYR-1 ISGIVLSLFANGKAQARPPATTPADTDAVRRCALYRNCFP-SMDVAYPAL
. : .* . . : . : : ..
MMAR_4104|M.marinum_M AFLCSEIARWVNEEGLSIDVSTGGELAVALHANFPPERITLHGNNKSVAE
MUL_3971|M.ulcerans_Agy99 AFLCSEIARWVNEEGLSIDVSTGGELAVALHANFPPERITLHGNNKSVAE
Mb1325|M.bovis_AF2122/97 AFLCSEVARWISEEGLCLDVCTGGELAVALHASFPPERITLHGNNKSVSE
Rv1293|M.tuberculosis_H37Rv AFLCSEVARWISEEGLCLDVCTGGELAVALHASFPPERITLHGNNKSVSE
MLBr_01128|M.leprae_Br4923 AFLCTEIARWIDEEGLSLDVCSGGELAVALHASFPPERISLHGNNKSVAE
Mflv_2297|M.gilvum_PYR-GCK AFLCGEIARWVDEEGLSLDVATGGELAVALHAGFPAERITVHGNNKSVAE
TH_2005|M.thermoresistible__bu AFLCTEIARWIDEEGLCLDVASGGELAVALHAGFPAERITLHGNNKSVEE
MAB_1156c|M.abscessus_ATCC_199 ALLTADIARWVAQEGLSLDVCSSGELATALAGGFDPERIIMHGNAKTPDE
MAV_1160|M.avium_104 SLLSTAVARWAREERLGVDVCSAGELATALAGGVDPARIVMHGNAKSPEE
MSMEG_5389|M.smegmatis_MC2_155 ALRDKAVARWVRDRSVAVEVRDGHELAIAQSAGVHPSRVTAHAEGLSIDE
Mvan_2486|M.vanbaalenii_PYR-1 VVRDHTVAKWVRAHDVAVHVRTSEELGAVISAGVHPARLTVHADGVLTNE
. :*:* . : :.* . **. . ... . *: *.: *
MMAR_4104|M.marinum_M LT-TAVKAGVGHIVLDSMIEIERLDAIAGEAGIVQDVFVRLTVGVEAHTH
MUL_3971|M.ulcerans_Agy99 LT-TAVKAGVGHIVLDSMIEIERLDAIAGEAGIVQDVFVRLTVGVEAHTH
Mb1325|M.bovis_AF2122/97 LT-AAVKAGVGHIVVDSMTEIERLDAIAGEAGIVQDVLVRLTVGVEAHTH
Rv1293|M.tuberculosis_H37Rv LT-AAVKAGVGHIVVDSMTEIERLDAIAGEAGIVQDVLVRLTVGVEAHTH
MLBr_01128|M.leprae_Br4923 LK-DAVKAGVGYIVLDSTTEIERLDAIAGEAGIVQDVLVRLTVGVEAHTH
Mflv_2297|M.gilvum_PYR-GCK LT-AAVKHGVGHVVVDSMIEIERLDRIAADNGVVQDVLVRVTVGVEAHTH
TH_2005|M.thermoresistible__bu LT-TAVEAGVGHVVLDSMIEIERLDRIAAAAGKVQDVLVRVTVGVEAHTH
MAB_1156c|M.abscessus_ATCC_199 LS-DAAAVGVGRIVVDSYTEIMFLATRLAKR---QRVLVRVTPDIDIHGH
MAV_1160|M.avium_104 LR-EAVRVGVGRIVLDSTIEIAYLAGLARRR---QPVLIRVTPDIDIHGH
MSMEG_5389|M.smegmatis_MC2_155 LVFCSVGLRVGRVVVTRADHVDVL--AVTR--RRQAVLLGSLR-------
Mvan_2486|M.vanbaalenii_PYR-1 LVFCAASLAVGHIVVNSVDQIEILTRAIGRQ-RRQAVLVRIVD-------
* :. ** :*: .: * * *::
MMAR_4104|M.marinum_M EFISTAHEDQKFGLSISSGAAMAAVRRVFATDHLRLVGLHSHIGSQIFDV
MUL_3971|M.ulcerans_Agy99 EFISTAHEDQKFGLSISSGAAMAAVRRVFATDHLRLVGLHSHIGSQIFDV
Mb1325|M.bovis_AF2122/97 EFISTAHEDQKFGLSVASGAAMAAVRRVFATDHLRLVGLHSHIGSQIFDV
Rv1293|M.tuberculosis_H37Rv EFISTAHEDQKFGLSVASGAAMAAVRRVFATDHLRLVGLHSHIGSQIFDV
MLBr_01128|M.leprae_Br4923 EFIATAHEDQKFGLSVASGAAMAAVRRVFATDNLRLVGLHSHIGSQIFDV
Mflv_2297|M.gilvum_PYR-GCK EFISTAHEDQKFGLSLASGAAMEAVRRVFEAGNLRLVGLHSHIGSQIFDV
TH_2005|M.thermoresistible__bu EFISTAHEDQKFGLSLASGAAMAAVRRVFATDNLRLVGLHSHIGSQIFDV
MAB_1156c|M.abscessus_ATCC_199 AAVTTGVTDQKFGFPLHEGHAIEAARRVLDQPLLNLVGLHCHIGSQVTDC
MAV_1160|M.avium_104 RAVTTGVSDQKFGFTLAGDHAADAVRRVLAHPILDLVGLHCHLGSQVSDP
MSMEG_5389|M.smegmatis_MC2_155 --------------RRLS-------RTVFDCATVDVVGMYGDVDHAAHHS
Mvan_2486|M.vanbaalenii_PYR-1 --------------TAANGESYAAIRAVLAGARLSLIGLHADIGSAAADF
* *: : ::*:: .:. .
MMAR_4104|M.marinum_M DGFELAAHRVIGLLREVVAEFGSEKTAQLATVDLGGGLGISYLSSDDPPP
MUL_3971|M.ulcerans_Agy99 DGFELAAHRVIGLLREVVAEFGSEKTAQLAMVDLGGGLGISYLSSDDPPP
Mb1325|M.bovis_AF2122/97 DGFELAAHRVIGLLRDVVGEFGPEKTAQIATVDLGGGLGISYLPSDDPPP
Rv1293|M.tuberculosis_H37Rv DGFELAAHRVIGLLRDVVGEFGPEKTAQIATVDLGGGLGISYLPSDDPPP
MLBr_01128|M.leprae_Br4923 AGFELAAHRVIGLLCDIVGEFDPEKTAQLSIVDLGGGLGISYLPDDDPPP
Mflv_2297|M.gilvum_PYR-GCK AGFEIAAHRVIGLLRDVVAEFGVEKTSQMDIVDLGGGLGISYLPDDNPPP
TH_2005|M.thermoresistible__bu AGFELAAHRVIGLLHDVVEEFGVEKTAQISTVDLGGGLGISYLPDDDPPP
MAB_1156c|M.abscessus_ATCC_199 ALYGETIRRMIAAMADIRARHG----VILGELNIGGGHGVPYRSGDPELD
MAV_1160|M.avium_104 ALYGEAIRRMIAAMADVRARHG----VILTELNIGGGHAIPYASGDPELN
MSMEG_5389|M.smegmatis_MC2_155 VSYPSAVGAMLAEMSRIRRDHG----VMLTRIALGG---RHLTFGAGAGE
Mvan_2486|M.vanbaalenii_PYR-1 VSYPAAVGDLICTMHRFRQSHG----RVLTRLSLGG---VPAPADDWVAE
: : :: : . .. : : :** .
MMAR_4104|M.marinum_M VGELADKLGAIVSNESAAVGLPAPRLVVEPGRAIAGPGTITLYEVGTVKD
MUL_3971|M.ulcerans_Agy99 VGELADKLGAIVSNESAAVGLPAPRLVVEPGRAIAGPGTITLYEVGTVKD
Mb1325|M.bovis_AF2122/97 IAELAAKLGTIVSDESTAVGLPTPKLVVEPGRAIAGPGTITLYEVGTVKD
Rv1293|M.tuberculosis_H37Rv IAELAAKLGTIVSDESTAVGLPTPKLVVEPGRAIAGPGTITLYEVGTVKD
MLBr_01128|M.leprae_Br4923 IFELAAKLGAIVSNESAAVGLPVPKLMVEPGRAIAGPGTITLYEVGTIKD
Mflv_2297|M.gilvum_PYR-GCK MSELAAKLRAIVARESAAVGLPAPKLVVEPGRAIAGPGTITLYEVGTVKD
TH_2005|M.thermoresistible__bu VEELAAKLKAIVRSESAAVGLPTPRLVVEPGRAIAGPGTITLYRVGTVKD
MAB_1156c|M.abscessus_ATCC_199 LAELRDFIDDALDAACAAERFPRPRVVVEPGRAISARAGVTLYRVVSVK-
MAV_1160|M.avium_104 LDQLADVIEDALDEACAAERFPRPQIVVEPGRAISGRAGVTLYRVCSVK-
MSMEG_5389|M.smegmatis_MC2_155 LSEVSAAVDGTLDDACATLRFPRPSVVVSASPAV----------------
Mvan_2486|M.vanbaalenii_PYR-1 LSRQAAVIDQSLDDACLTVDFPRPTVTVSAFTVASGKRAA----------
: . : : . : :* * : *.. .
MMAR_4104|M.marinum_M VDVSATANRRYVSVDGGMSDNIRTALYDAQYDVRLVSRVSDAPATLARVV
MUL_3971|M.ulcerans_Agy99 VDVSATANRRYVSVDGGMSDNIRTALYDAQYDVRLVSRVSDAPAMLARVV
Mb1325|M.bovis_AF2122/97 VDVSATAHRRYVSVDGGMSDNIRTALYGAQYDVRLVSRVSDAPPVPARLV
Rv1293|M.tuberculosis_H37Rv VDVSATAHRRYVSVDGGMSDNIRTALYGAQYDVRLVSRVSDAPPVPARLV
MLBr_01128|M.leprae_Br4923 VDVSATAHRRYVSIDGGMSDNIRTALYDAQYDVRLVSRTSDAPAAPASIV
Mflv_2297|M.gilvum_PYR-GCK VAVSPTSHRRYVSVDGGMSDNIRTSLYGAEYDVRLLSRVSDATPTLSRVV
TH_2005|M.thermoresistible__bu VAISPTAYRRYVSVDGGMSDNIRTALYGAQYDARLVSRRTDAEPALGRIV
MAB_1156c|M.abscessus_ATCC_199 ---TQPGGRTFVAVDGGMSDNPRVSLYSAKYSVALANRHPSAPPQLVTVA
MAV_1160|M.avium_104 ---TQCGGRTFVAVDGGMSDNPRVSLYGARYAVALANRHPLGLKQRVTVA
MSMEG_5389|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_2486|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4104|M.marinum_M GKHCESGDIIVRDTWVPDDLQPGDLVAVAATGAYCYSLSSRYNMIGRPAV
MUL_3971|M.ulcerans_Agy99 GKHCESGDIIVRDTWVPDDLQPGDLVAVAATGAYCYSLSSRYNMIGRPAV
Mb1325|M.bovis_AF2122/97 GKHCESGDIIVRDTWVPDDIRPGDLVAVAATGAYCYSLSSRYNMVGRPAV
Rv1293|M.tuberculosis_H37Rv GKHCESGDIIVRDTWVPDDIRPGDLVAVAATGAYCYSLSSRYNMVGRPAV
MLBr_01128|M.leprae_Br4923 GKHCESGDIVVRDTWVPDDLKPGDLVGVAATGAYCYSLSSRYNMLGRPAV
Mflv_2297|M.gilvum_PYR-GCK GKHCESGDIVVRDAWLSEDVRPGDLLGVAATGAYCYSMSSRYNLIGRPAV
TH_2005|M.thermoresistible__bu GKHCESGDIVVRDVWVPGDVQPGDLIGVAATGAYCYSMSSRYNLIGRPAV
MAB_1156c|M.abscessus_ATCC_199 GRHCESGDEIARDVSLPSDVRPGDVLAVACTGAYHHSMASSYNLVGRPPV
MAV_1160|M.avium_104 GRHCESGDEIARDVGLPADLRPGDLLAVACTGAYHHSMASNYNMVGRPPL
MSMEG_5389|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_2486|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4104|M.marinum_M VAVRAGKARLVLRRETVDDLLSLEVR--
MUL_3971|M.ulcerans_Agy99 VAVRAGKARLVLRRETVDDLLSLEVR--
Mb1325|M.bovis_AF2122/97 VAVHAGNARLVLRRETVDDLLSLEVR--
Rv1293|M.tuberculosis_H37Rv VAVHAGNARLVLRRETVDDLLSLEVR--
MLBr_01128|M.leprae_Br4923 VAVCAGQARLILRRETVDDLLSLEVR--
Mflv_2297|M.gilvum_PYR-GCK VAVRDGRARLILRRETVEDLLSLEVR--
TH_2005|M.thermoresistible__bu VVVRDGRARLILRRETVDDLLSLEVR--
MAB_1156c|M.abscessus_ATCC_199 VAVRDGRVRELVRRETIADLLSRDRGAR
MAV_1160|M.avium_104 VAVSEGRARELVRRETVADLLARDRG--
MSMEG_5389|M.smegmatis_MC2_155 ----------------------------
Mvan_2486|M.vanbaalenii_PYR-1 ----------------------------