For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LDILPSLGHAAPPRFDPAIWPVTAHPDEEGRLCVGGVPLGDVADEFGTPVYVVDEADFRCRARRYRRALR GVEVVYAGKSLLSTAVARWAREERLGVDVCSAGELATALAGGVDPARIVMHGNAKSPEELREAVRVGVGR IVLDSTIEIAYLAGLARRRQPVLIRVTPDIDIHGHRAVTTGVSDQKFGFTLAGDHAADAVRRVLAHPILD LVGLHCHLGSQVSDPALYGEAIRRMIAAMADVRARHGVILTELNIGGGHAIPYASGDPELNLDQLADVIE DALDEACAAERFPRPQIVVEPGRAISGRAGVTLYRVCSVKTQCGGRTFVAVDGGMSDNPRVSLYGARYAV ALANRHPLGLKQRVTVAGRHCESGDEIARDVGLPADLRPGDLLAVACTGAYHHSMASNYNMVGRPPLVAV SEGRARELVRRETVADLLARDRG*
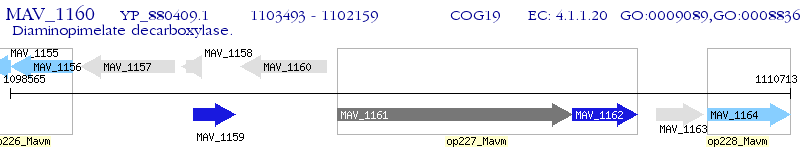
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1160 | lysA | - | 100% (444) | diaminopimelate decarboxylase |
| M. avium 104 | MAV_1508 | lysA | 3e-94 | 45.06% (435) | diaminopimelate decarboxylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1325 | lysA | 6e-98 | 46.21% (435) | diaminopimelate decarboxylase LysA |
| M. gilvum PYR-GCK | Mflv_2297 | - | 1e-100 | 47.26% (438) | diaminopimelate decarboxylase |
| M. tuberculosis H37Rv | Rv1293 | lysA | 6e-98 | 46.21% (435) | diaminopimelate decarboxylase LysA |
| M. leprae Br4923 | MLBr_01128 | lysA | 7e-97 | 44.79% (451) | diaminopimelate decarboxylase |
| M. abscessus ATCC 19977 | MAB_1156c | - | 0.0 | 74.10% (444) | diaminopimelate decarboxylase LysA |
| M. marinum M | MMAR_4104 | lysA | 4e-97 | 46.21% (435) | diaminopimelate decarboxylase LysA |
| M. smegmatis MC2 155 | MSMEG_4958 | lysA | 3e-96 | 45.75% (435) | diaminopimelate decarboxylase |
| M. thermoresistible (build 8) | TH_2005 | lysA | 1e-100 | 46.90% (435) | PROBABLE DIAMINOPIMELATE DECARBOXYLASE LYSA (DAP |
| M. ulcerans Agy99 | MUL_3971 | lysA | 2e-97 | 46.21% (435) | diaminopimelate decarboxylase LysA |
| M. vanbaalenii PYR-1 | Mvan_4349 | - | 3e-97 | 47.36% (435) | diaminopimelate decarboxylase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2297|M.gilvum_PYR-GCK MIAHPAGPRHAEEVPSAAAPERPQSAADVSRLAPNVWPQNTVRGDDGAVS
Mvan_4349|M.vanbaalenii_PYR-1 MIAHPAGPRHAEEVHPVATPDRPRTAAEVLQLAPNVWPQNAVRGADGVVS
MSMEG_4958|M.smegmatis_MC2_155 MNAHPAGPRHAEELHHADVPDKPQSPDEIMLLAPNVWPRNLVRGADGVVT
TH_2005|M.thermoresistible__bu VIAHPAGPRHAEEIHHGETPPRPQSPDELLLLAPNVWPRNTVRESDGVVS
Mb1325|M.bovis_AF2122/97 -------------------------MNELLHLAPNVWPRNTTRDEVGVVC
Rv1293|M.tuberculosis_H37Rv -------------------------MNELLHLAPNVWPRNTTRDEVGVVC
MMAR_4104|M.marinum_M MNVHPAGPRHAEESRHPESPQRPQSAEELLYLAPNVWPRNTTRDEQGCAR
MUL_3971|M.ulcerans_Agy99 MNVHPAGPRHAEESRHPESPQRPQSAEELLYLAPNVWPRNTARDEQGCAR
MLBr_01128|M.leprae_Br4923 MNVHTAGPRHAEKTRHTATPQRVQPSDDLLRLASNVWPRNITRDETGVAC
MAV_1160|M.avium_104 --MLDILP----SLGHAAPP----------RFDPAIWPVTAHPDEEGRLC
MAB_1156c|M.abscessus_ATCC_199 MTLLDLLP----SLHNVARP----------RVDPAVWPVTTHLDEQGRMC
. . :** . *
Mflv_2297|M.gilvum_PYR-GCK IAGVPVADIAAEFGTPVFVIDEADFRSRCRDISSAFGGGEYVHYAAKAFL
Mvan_4349|M.vanbaalenii_PYR-1 IAGVSVADIAAEFGTPVFVIDEADFRARCRDIAAAFGGGEHVRYAAKAFL
MSMEG_4958|M.smegmatis_MC2_155 IAGVPVTDLAAKFGTPLFVIDEDDFRTRCRDIAAAFGGGQYVHYAAKAFL
TH_2005|M.thermoresistible__bu IAGVRVTDIADEYGTPTFVIDEDDFRSRCREMAAAFGGGENVHYAAKAFL
Mb1325|M.bovis_AF2122/97 IAGIPLTQLAQEYGTPLFVIDEDDFRSRCRETAAAFGSGANVHYAAKAFL
Rv1293|M.tuberculosis_H37Rv IAGIPLTQLAQEYGTPLFVIDEDDFRSRCRETAAAFGSGANVHYAAKAFL
MMAR_4104|M.marinum_M IAGVPLTELAREYGTPLFVIDEDDFRSRCQEIAAAFGGGHNVNYAAKAFL
MUL_3971|M.ulcerans_Agy99 IAGVPLTELAREYGTPLFVIDEDDFRSRCQEIAAAFGGGHNVNYAAKAFL
MLBr_01128|M.leprae_Br4923 IAGNKLTDLAGEYGTPLFVIDEDDFRFRCREIAAAFGGGENVHYAAKAFL
MAV_1160|M.avium_104 VGGVPLGDVADEFGTPVYVVDEADFRCRARRYRRALR-GVEVVYAGKSLL
MAB_1156c|M.abscessus_ATCC_199 VGETALSEIADQFRTPAYVLDEEDFRHRARRYRSTLR-EIEVAYAGKALL
:. : ::* :: ** :*:** *** *.: :: * **.*::*
Mflv_2297|M.gilvum_PYR-GCK CGEIARWVDEEGLSLDVATGGELAVALHAGFPAERITVHGNNKSVAELTA
Mvan_4349|M.vanbaalenii_PYR-1 CTEVARWIDEEGLSLDVASGGELAVALHAGFPAERIALHGNNKSVAELTA
MSMEG_4958|M.smegmatis_MC2_155 CSEIARWVDEEGLSLDVATGGELAVALHAGFPASRITVHGNNKSVAELTA
TH_2005|M.thermoresistible__bu CTEIARWIDEEGLCLDVASGGELAVALHAGFPAERITLHGNNKSVEELTT
Mb1325|M.bovis_AF2122/97 CSEVARWISEEGLCLDVCTGGELAVALHASFPPERITLHGNNKSVSELTA
Rv1293|M.tuberculosis_H37Rv CSEVARWISEEGLCLDVCTGGELAVALHASFPPERITLHGNNKSVSELTA
MMAR_4104|M.marinum_M CSEIARWVNEEGLSIDVSTGGELAVALHANFPPERITLHGNNKSVAELTT
MUL_3971|M.ulcerans_Agy99 CSEIARWVNEEGLSIDVSTGGELAVALHANFPPERITLHGNNKSVAELTT
MLBr_01128|M.leprae_Br4923 CTEIARWIDEEGLSLDVCSGGELAVALHASFPPERISLHGNNKSVAELKD
MAV_1160|M.avium_104 STAVARWAREERLGVDVCSAGELATALAGGVDPARIVMHGNAKSPEELRE
MAB_1156c|M.abscessus_ATCC_199 TADIARWVAQEGLSLDVCSSGELATALAGGFDPERIIMHGNAKTPDELSD
:*** :* * :**.:.****.** ... . ** :*** *: **
Mflv_2297|M.gilvum_PYR-GCK AVKHGVGHVVVDSMIEIERLDRIAADNGVVQDVLVRVTVGVEAHTHEFIS
Mvan_4349|M.vanbaalenii_PYR-1 AVEAGIEHVVVDSMTEIDRLDKIAGEAGVVQDVLIRVTVGVEAHTHEFIS
MSMEG_4958|M.smegmatis_MC2_155 AVQAGVGHVVLDSEIEIERLDQIAGAAGVVQDVLVRVTVGVEAHTHEFIS
TH_2005|M.thermoresistible__bu AVEAGVGHVVLDSMIEIERLDRIAAAAGKVQDVLVRVTVGVEAHTHEFIS
Mb1325|M.bovis_AF2122/97 AVKAGVGHIVVDSMTEIERLDAIAGEAGIVQDVLVRLTVGVEAHTHEFIS
Rv1293|M.tuberculosis_H37Rv AVKAGVGHIVVDSMTEIERLDAIAGEAGIVQDVLVRLTVGVEAHTHEFIS
MMAR_4104|M.marinum_M AVKAGVGHIVLDSMIEIERLDAIAGEAGIVQDVFVRLTVGVEAHTHEFIS
MUL_3971|M.ulcerans_Agy99 AVKAGVGHIVLDSMIEIERLDAIAGEAGIVQDVFVRLTVGVEAHTHEFIS
MLBr_01128|M.leprae_Br4923 AVKAGVGYIVLDSTTEIERLDAIAGEAGIVQDVLVRLTVGVEAHTHEFIA
MAV_1160|M.avium_104 AVRVGVGRIVLDSTIEIAYLAGLARRR---QPVLIRVTPDIDIHGHRAVT
MAB_1156c|M.abscessus_ATCC_199 AAAVGVGRIVVDSYTEIMFLATRLAKR---QRVLVRVTPDIDIHGHAAVT
*. *: :*:** ** * * *::*:* .:: * * ::
Mflv_2297|M.gilvum_PYR-GCK TAHEDQKFGLSLASGAAMEAVRRVFEAGNLRLVGLHSHIGSQIFDVAGFE
Mvan_4349|M.vanbaalenii_PYR-1 TAHEDQKFGLSLASGAAMTAVRRVFETDHLRLVGLHSHIGSQIFDVAGFE
MSMEG_4958|M.smegmatis_MC2_155 TAHEDQKFGLSLATGAAMNAIRRVFATDNLRLVGLHSHIGSQIFDVAGFE
TH_2005|M.thermoresistible__bu TAHEDQKFGLSLASGAAMAAVRRVFATDNLRLVGLHSHIGSQIFDVAGFE
Mb1325|M.bovis_AF2122/97 TAHEDQKFGLSVASGAAMAAVRRVFATDHLRLVGLHSHIGSQIFDVDGFE
Rv1293|M.tuberculosis_H37Rv TAHEDQKFGLSVASGAAMAAVRRVFATDHLRLVGLHSHIGSQIFDVDGFE
MMAR_4104|M.marinum_M TAHEDQKFGLSISSGAAMAAVRRVFATDHLRLVGLHSHIGSQIFDVDGFE
MUL_3971|M.ulcerans_Agy99 TAHEDQKFGLSISSGAAMAAVRRVFATDHLRLVGLHSHIGSQIFDVDGFE
MLBr_01128|M.leprae_Br4923 TAHEDQKFGLSVASGAAMAAVRRVFATDNLRLVGLHSHIGSQIFDVAGFE
MAV_1160|M.avium_104 TGVSDQKFGFTLAGDHAADAVRRVLAHPILDLVGLHCHLGSQVSDPALYG
MAB_1156c|M.abscessus_ATCC_199 TGVTDQKFGFPLHEGHAIEAARRVLDQPLLNLVGLHCHIGSQVTDCALYG
*. *****:.: . * * ***: * *****.*:***: * :
Mflv_2297|M.gilvum_PYR-GCK IAAHRVIGLLRDVVAEFGVEKTSQMDIVDLGGGLGISYLPDDNPPPMSEL
Mvan_4349|M.vanbaalenii_PYR-1 LAAHRVIGLLRDAVAEFGVEKTSQMSIVDLGGGLGISYLPHDDPPPMGEL
MSMEG_4958|M.smegmatis_MC2_155 IAAHRVIGLLRDVVAEFGVEKTSQMSIVDLGGGLGISYLPHDDPPPMKEL
TH_2005|M.thermoresistible__bu LAAHRVIGLLHDVVEEFGVEKTAQISTVDLGGGLGISYLPDDDPPPVEEL
Mb1325|M.bovis_AF2122/97 LAAHRVIGLLRDVVGEFGPEKTAQIATVDLGGGLGISYLPSDDPPPIAEL
Rv1293|M.tuberculosis_H37Rv LAAHRVIGLLRDVVGEFGPEKTAQIATVDLGGGLGISYLPSDDPPPIAEL
MMAR_4104|M.marinum_M LAAHRVIGLLREVVAEFGSEKTAQLATVDLGGGLGISYLSSDDPPPVGEL
MUL_3971|M.ulcerans_Agy99 LAAHRVIGLLREVVAEFGSEKTAQLAMVDLGGGLGISYLSSDDPPPVGEL
MLBr_01128|M.leprae_Br4923 LAAHRVIGLLCDIVGEFDPEKTAQLSIVDLGGGLGISYLPDDDPPPIFEL
MAV_1160|M.avium_104 EAIRRMIAAMADVRARHG----VILTELNIGGGHAIPYASGDPELNLDQL
MAB_1156c|M.abscessus_ATCC_199 ETIRRMIAAMADIRARHG----VILGELNIGGGHGVPYRSGDPELDLAEL
: :*:*. : : ... : :::*** .:.* . * : :*
Mflv_2297|M.gilvum_PYR-GCK AAKLRAIVARESAAVGLPAPKLVVEPGRAIAGPGTITLYEVGTVKDVAVS
Mvan_4349|M.vanbaalenii_PYR-1 ADKLHAIVRHESAAVGLPAPRLVVEPGRAIAGPGTITLYEVGTVKDVAIS
MSMEG_4958|M.smegmatis_MC2_155 ADKLLEIVRTESAAVGLPTPKLVVEPGRAIAGPGTITLYEVGTVKDVAVS
TH_2005|M.thermoresistible__bu AAKLKAIVRSESAAVGLPTPRLVVEPGRAIAGPGTITLYRVGTVKDVAIS
Mb1325|M.bovis_AF2122/97 AAKLGTIVSDESTAVGLPTPKLVVEPGRAIAGPGTITLYEVGTVKDVDVS
Rv1293|M.tuberculosis_H37Rv AAKLGTIVSDESTAVGLPTPKLVVEPGRAIAGPGTITLYEVGTVKDVDVS
MMAR_4104|M.marinum_M ADKLGAIVSNESAAVGLPAPRLVVEPGRAIAGPGTITLYEVGTVKDVDVS
MUL_3971|M.ulcerans_Agy99 ADKLGAIVSNESAAVGLPAPRLVVEPGRAIAGPGTITLYEVGTVKDVDVS
MLBr_01128|M.leprae_Br4923 AAKLGAIVSNESAAVGLPVPKLMVEPGRAIAGPGTITLYEVGTIKDVDVS
MAV_1160|M.avium_104 ADVIEDALDEACAAERFPRPQIVVEPGRAISGRAGVTLYRVCSVK----T
MAB_1156c|M.abscessus_ATCC_199 RDFIDDALDAACAAERFPRPRVVVEPGRAISARAGVTLYRVVSVK----T
: : .:* :* *:::*******:. . :***.* ::* :
Mflv_2297|M.gilvum_PYR-GCK PTSHRRYVSVDGGMSDNIRTSLYGAEYDVRLLSRVSDATPTLSRVVGKHC
Mvan_4349|M.vanbaalenii_PYR-1 SDRNRRYVSVDGGMSDNIRTSLYDAEYDVRLVSRTSNAEPTLSRVVGKHC
MSMEG_4958|M.smegmatis_MC2_155 QTAHRRYVSVDGGMSDNIRTSLYAAQYDARLVSRVSDAPPALARIVGKHC
TH_2005|M.thermoresistible__bu PTAYRRYVSVDGGMSDNIRTALYGAQYDARLVSRRTDAEPALGRIVGKHC
Mb1325|M.bovis_AF2122/97 ATAHRRYVSVDGGMSDNIRTALYGAQYDVRLVSRVSDAPPVPARLVGKHC
Rv1293|M.tuberculosis_H37Rv ATAHRRYVSVDGGMSDNIRTALYGAQYDVRLVSRVSDAPPVPARLVGKHC
MMAR_4104|M.marinum_M ATANRRYVSVDGGMSDNIRTALYDAQYDVRLVSRVSDAPATLARVVGKHC
MUL_3971|M.ulcerans_Agy99 ATANRRYVSVDGGMSDNIRTALYDAQYDVRLVSRVSDAPAMLARVVGKHC
MLBr_01128|M.leprae_Br4923 ATAHRRYVSIDGGMSDNIRTALYDAQYDVRLVSRTSDAPAAPASIVGKHC
MAV_1160|M.avium_104 QCGGRTFVAVDGGMSDNPRVSLYGARYAVALANRHPLGLKQRVTVAGRHC
MAB_1156c|M.abscessus_ATCC_199 QPGGRTFVAVDGGMSDNPRVSLYSAKYSVALANRHPSAPPQLVTVAGRHC
* :*::******* *.:** *.* . * .* . . :.*:**
Mflv_2297|M.gilvum_PYR-GCK ESGDIVVRDAWLSEDVRPGDLLGVAATGAYCYSMSSRYNLIGRPAVVAVR
Mvan_4349|M.vanbaalenii_PYR-1 ESGDIVVRDAWIPDDVGPGDLLGVAATGAYCYSMSSRYNLIGRPAVVAVR
MSMEG_4958|M.smegmatis_MC2_155 ESGDIVVRDTWVSDDIAPGDLIGVAATGAYCYSMSSRYNLLCRPAVVAVA
TH_2005|M.thermoresistible__bu ESGDIVVRDVWVPGDVQPGDLIGVAATGAYCYSMSSRYNLIGRPAVVVVR
Mb1325|M.bovis_AF2122/97 ESGDIIVRDTWVPDDIRPGDLVAVAATGAYCYSLSSRYNMVGRPAVVAVH
Rv1293|M.tuberculosis_H37Rv ESGDIIVRDTWVPDDIRPGDLVAVAATGAYCYSLSSRYNMVGRPAVVAVH
MMAR_4104|M.marinum_M ESGDIIVRDTWVPDDLQPGDLVAVAATGAYCYSLSSRYNMIGRPAVVAVR
MUL_3971|M.ulcerans_Agy99 ESGDIIVRDTWVPDDLQPGDLVAVAATGAYCYSLSSRYNMIGRPAVVAVR
MLBr_01128|M.leprae_Br4923 ESGDIVVRDTWVPDDLKPGDLVGVAATGAYCYSLSSRYNMLGRPAVVAVC
MAV_1160|M.avium_104 ESGDEIARDVGLPADLRPGDLLAVACTGAYHHSMASNYNMVGRPPLVAVS
MAB_1156c|M.abscessus_ATCC_199 ESGDEIARDVSLPSDVRPGDVLAVACTGAYHHSMASSYNLVGRPPVVAVR
**** :.**. :. *: ***::.**.**** :*::* **:: **.:*.*
Mflv_2297|M.gilvum_PYR-GCK DGRARLILRRETVEDLLSLEVR----
Mvan_4349|M.vanbaalenii_PYR-1 DGRARLILRRETFDDLLSLEVSEVKR
MSMEG_4958|M.smegmatis_MC2_155 DGQARLVLRRETVEDLLSLEVKGQ--
TH_2005|M.thermoresistible__bu DGRARLILRRETVDDLLSLEVR----
Mb1325|M.bovis_AF2122/97 AGNARLVLRRETVDDLLSLEVR----
Rv1293|M.tuberculosis_H37Rv AGNARLVLRRETVDDLLSLEVR----
MMAR_4104|M.marinum_M AGKARLVLRRETVDDLLSLEVR----
MUL_3971|M.ulcerans_Agy99 AGKARLVLRRETVDDLLSLEVR----
MLBr_01128|M.leprae_Br4923 AGQARLILRRETVDDLLSLEVR----
MAV_1160|M.avium_104 EGRARELVRRETVADLLARDRG----
MAB_1156c|M.abscessus_ATCC_199 DGRVRELVRRETIADLLSRDRGAR--
*..* ::****. ***: :