For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MIALTIVLAVFVGIALGMLGGGGSILTVPLLAYVAGMDAKQAVTTSLLVVGVTSAVSAVSHARAGRVQWR AGLSFGAAGMAGAYAGGVLGRFIPGTALLIAFAVVMVAAAIPMLRGRRAPEATTAHGLPVVRVVSLGLAV GGLTGMIGAGGGFLVVPALTLLGGLPMPAAVGTSLIVIALNSFSGLAGHLTGLQIDWALAAAVTGAAVAG CLIGTRLITKINADALRKAFGWFVLAMSSVILAQEIHPFVGLAAASLTVIAASMTMVCARFSRCPLRRVT RLSMARPAVA
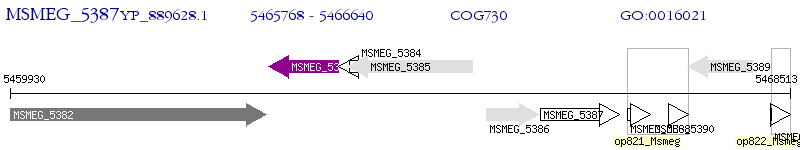
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5387 | - | - | 100% (290) | hypothetical protein MSMEG_5387 |
| M. smegmatis MC2 155 | MSMEG_3735 | - | 4e-11 | 25.87% (286) | YtnM protein |
| M. smegmatis MC2 155 | MSMEG_2287 | - | 2e-07 | 26.53% (245) | transport protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2902c | - | 1e-05 | 28.75% (240) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_0983 | - | 1e-115 | 71.82% (291) | hypothetical protein Mflv_0983 |
| M. tuberculosis H37Rv | Rv2877c | - | 1e-05 | 28.75% (240) | integral membrane protein |
| M. leprae Br4923 | MLBr_00468 | - | 9e-09 | 23.65% (241) | hypothetical protein MLBr_00468 |
| M. abscessus ATCC 19977 | MAB_1170 | - | 7e-21 | 30.97% (268) | hypothetical protein MAB_1170 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3519 | - | 5e-07 | 26.14% (241) | hypothetical protein MAV_3519 |
| M. thermoresistible (build 8) | TH_4293 | - | 1e-111 | 69.28% (293) | PUTATIVE putative protein of unknown function DUF81 |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1828 | - | 1e-112 | 72.07% (290) | hypothetical protein Mvan_1828 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0983|M.gilvum_PYR-GCK -----MMALTVGLAVFVGIALGLLGGGGSILTVPLLAYVAG--MDAKQAI
Mvan_1828|M.vanbaalenii_PYR-1 -----MIALTIGLAVFVGIALGLLGGGGSILTVPLLAYVAG--MDAKAAI
TH_4293|M.thermoresistible__bu -----MIGLIIALAVLVGVALGLLGGGGSILMVPLLAYVAG--MDAKAAI
MSMEG_5387|M.smegmatis_MC2_155 -----MIALTIVLAVFVGIALGMLGGGGSILTVPLLAYVAG--MDAKQAV
MLBr_00468|M.leprae_Br4923 --MLVSHLATIGLAGFGAGAINALVGSGTLITFPTLVALGYSPVTSTMSN
MAV_3519|M.avium_104 -------MFVICLAGLAAGAINALVGSGTLITFPTLVALGYPPVTATMSN
Mb2902c|M.bovis_AF2122/97 -----MNEALIGLAFAAGLVAALNPCGFAMLPAYLLLVVHGQ-DSAGRTG
Rv2877c|M.tuberculosis_H37Rv -----MNEALIGLAFAAGLVAALNPCGFAMLPAYLLLVVYGQ-DSAGRTG
MAB_1170|M.abscessus_ATCC_1997 MTERPGVGTLARIGVLGGMSGGLLGGGTGVITVPSLARATT---LSRAVI
:. . . :: * :
Mflv_0983|M.gilvum_PYR-GCK ATSLLVVGVTSAIG--AVSHARAGRVQWRTGLIFGAAGMAG----AYAGG
Mvan_1828|M.vanbaalenii_PYR-1 ATSLLVVGVTSAIG--AISHARAGRVQWRTGLIFGAAGMAG----AYAGG
TH_4293|M.thermoresistible__bu ATSLLVVGVTSAVG--AVSHARAGQVRWRTGLIFGAAGMAG----AYAGG
MSMEG_5387|M.smegmatis_MC2_155 TTSLLVVGVTSAVS--AVSHARAGRVQWRAGLSFGAAGMAG----AYAGG
MLBr_00468|M.leprae_Br4923 AVGLVAGGVSATWGYRAELGDRWSRLRWQIPMSLTGAALGAFLLLHLPEK
MAV_3519|M.avium_104 ATGLVAGSVSGTWGYRAELRGQWDRLRWQLPASLAGAGLGAYLLLHLPEK
Mb2902c|M.bovis_AF2122/97 PLSAVGRAAAATVGMALGFLTVFGIFGALTISAATAVQRYLPYATVLIGL
Rv2877c|M.tuberculosis_H37Rv PLSAVGRAAAATVGMALGFLTVFGIFGALTISAATAVQRYLPYATVLIGL
MAB_1170|M.abscessus_ATCC_1997 HGTATLPNVSAAIAGSTVYALHGAKVDVVAGAGLMAGGVIG----AVVGA
.:.: . . .
Mflv_0983|M.gilvum_PYR-GCK LLARFIPGTVLLIGFALMMIATAVAMLR-------GRKNVQATEGS--HR
Mvan_1828|M.vanbaalenii_PYR-1 LLARYIPGTILLIGFAVMMIATAIAMLR-------GRKTIETGETG--HR
TH_4293|M.thermoresistible__bu ILARFIPGTVLLLGFAVMMAATAVAMLR-------GRRNTATPDTGGDHR
MSMEG_5387|M.smegmatis_MC2_155 VLGRFIPGTALLIAFAVVMVAAAIPMLR-------GRR---APEATTAHG
MLBr_00468|M.leprae_Br4923 VFTAIVP-MLLVLALVLVVAGPRIQSWT-------RCRAKAAVNRAEHTT
MAV_3519|M.avium_104 VFAEIVP-VLLVAALVLVVTGPRIQAWT-------ARRAELAGRSPEHVP
Mb2902c|M.bovis_AF2122/97 ALIALGGWLLLGRGLTALTPRSLGVRWA-------PTVRLGSMYGYGISY
Rv2877c|M.tuberculosis_H37Rv ALIALGGWLLLGRGLTALTPRSLGVRWA-------PTVRLGSMYGYGISY
MAB_1170|M.abscessus_ATCC_1997 KLVARIPEWILKALFVFVLLATAAKLLLSAAGIDPSAGEALLPEGVLAHV
: * :. : .
Mflv_0983|M.gilvum_PYR-GCK LPVPKILAEG-LLVGLVTGLVGAGGGFLVVPALALLGGLPMP-VAVGTSL
Mvan_1828|M.vanbaalenii_PYR-1 LPVPKIVAEG-LIVGLVTGLVGAGGGFLVVPALALLGGLPMP-IAVGTSL
TH_4293|M.thermoresistible__bu LPVPRILADG-LVVGLVTGLVGAGGGFLVVPALALLGGLSMP-AAVGTSL
MSMEG_5387|M.smegmatis_MC2_155 LPVVRVVSLG-LAVGGLTGMIGAGGGFLVVPALTLLGGLPMP-AAVGTSL
MLBr_00468|M.leprae_Br4923 PAQMAMLALGTFAVSVYGGYFTAGQGILLVSLMSILLPESMQRINAAKNL
MAV_3519|M.avium_104 PGRMATLVFGTFAVGVYGGYFTAAQGILLVGLMGALLPESVQRMNAAKNL
Mb2902c|M.bovis_AF2122/97 AVASLSCTIGPFLAVTGAGLRGGSVVGSVAIYLAYVAGLTLVVGVLAVAA
Rv2877c|M.tuberculosis_H37Rv AVASLSCTIGPFLAVTGAGLRGGSVVGSVAIYLAYVAGLTLVVGVLAVAA
MAB_1170|M.abscessus_ATCC_1997 PSVIAIGAVVGFVVGAWSAALGLGGGLLTVPAMLVLFGSELH-VAEGTSL
: . . . . : : : .
Mflv_0983|M.gilvum_PYR-GCK IVIAMKSFAGLGGYLSSVQ-IDWSLALAVTAAAVVGALLGARLTSMVNPD
Mvan_1828|M.vanbaalenii_PYR-1 IVIAMKSFAGLAGYLTSVH-INWTLALAVTAAAVIGALIGARLTAMINPD
TH_4293|M.thermoresistible__bu VVIAMKSFAGLAGYLTSVQ-IDWPVALAVTVAAVAGALLGSRLTALVNPD
MSMEG_5387|M.smegmatis_MC2_155 IVIALNSFSGLAGHLTGLQ-IDWALAAAVTGAAVAGCLIGTRLITKINAD
MLBr_00468|M.leprae_Br4923 LSMLVNLTAAVGYTLVAFDRINWSAAGMIAASSLVGGLVGTRYARRLSPN
MAV_3519|M.avium_104 LALVVNVVAAVAYTSVAFGRISWAAAGLIAAGSLVGGLLGARYGRRLSGG
Mb2902c|M.bovis_AF2122/97 ATASSALADRLRRILPFVNRISGALLVVVGLYVGYYGLYELRLIAGVGAN
Rv2877c|M.tuberculosis_H37Rv ATASSALADRLRRILPFVNRISGALLVVVGLYVGYYGLYELRLIAGVGAN
MAB_1170|M.abscessus_ATCC_1997 MVMLPNALVAATTHLRQGT-ADAPIGFRLAAFALPGTVIGALLALALNAR
. . : : :.
Mflv_0983|M.gilvum_PYR-GCK SLRKAFGWFVLAMSSVILAQEIHLAVGIAGAALTAIAALMTFACSRYAHC
Mvan_1828|M.vanbaalenii_PYR-1 SLRKTFGWFVLVMSSVILAQEIHPAVGITAAALTLIAAATTLACQRYAHC
TH_4293|M.thermoresistible__bu TLRQAFGWFVLTMSAVILAQEAHPAVGAAAAVLTAVAAALTFTCSRYGRC
MSMEG_5387|M.smegmatis_MC2_155 ALRKAFGWFVLAMSSVILAQEIHPFVGLAAASLTVIAASMTMVCARFSRC
MLBr_00468|M.leprae_Br4923 VLRATIVAIGLIGLVRLLAV------------------------------
MAV_3519|M.avium_104 ALRAIIVVVGLIGLYRLLAVA-----------------------------
Mb2902c|M.bovis_AF2122/97 PQDAVIAAAGRLQGALAGWVNQHGAWPWAVLLVVLVVGAFAGTWFRRVRR
Rv2877c|M.tuberculosis_H37Rv PQDAVIAAAGRLQGALAGWVNQHGAWPWAVLLVVLVVGAFAGTWFRRVRR
MAB_1170|M.abscessus_ATCC_1997 WLALVFGSYLVFIAVREIVRAIRTAAAKTKKSGPAPVPQVCVGG------
:
Mflv_0983|M.gilvum_PYR-GCK PLRRITGIGAARGAHA
Mvan_1828|M.vanbaalenii_PYR-1 PLSRLTTRLTTPKAAA
TH_4293|M.thermoresistible__bu PLRRVAEALSSRPATA
MSMEG_5387|M.smegmatis_MC2_155 PLRRVTRLSMARPAVA
MLBr_00468|M.leprae_Br4923 ----------------
MAV_3519|M.avium_104 ----------------
Mb2902c|M.bovis_AF2122/97 ----------------
Rv2877c|M.tuberculosis_H37Rv ----------------
MAB_1170|M.abscessus_ATCC_1997 ----------------