For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TVWDVVLLVFAGIAGGLTGSIAGLASVATYPALLVVGLPPVAANVTNTVAVVFNGVGSIAGSRPELAGQG AWLKRIIPVAALGGVAGAALLLSTPAEGFEKIVPFLLGFASVAILLPRREHRSARVANHRDHLIRTGVEA AAIFLITIYGGYFGAAAGVLLLALMLRAGGATLPHANAGKNVILGVANLVASAIFVVFAPVYWPAVVPLG IGCLIGSRLGPIIVRHAPSTPLRWLIGVAGIALAIKLALDTY*
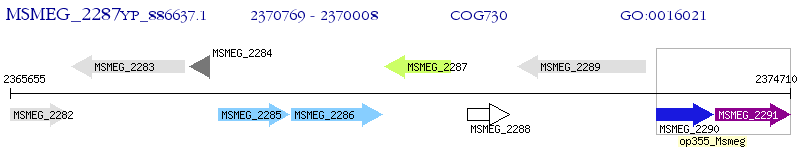
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2287 | - | - | 100% (253) | transport protein |
| M. smegmatis MC2 155 | MSMEG_1007 | - | e-140 | 100.00% (253) | transport protein |
| M. smegmatis MC2 155 | MSMEG_3737 | - | 3e-32 | 33.73% (255) | integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4317 | - | 2e-90 | 68.29% (246) | hypothetical protein Mflv_4317 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00468 | - | 2e-26 | 30.49% (246) | hypothetical protein MLBr_00468 |
| M. abscessus ATCC 19977 | MAB_3982 | - | 5e-29 | 33.20% (253) | hypothetical protein MAB_3982 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3519 | - | 4e-29 | 34.27% (248) | hypothetical protein MAV_3519 |
| M. thermoresistible (build 8) | TH_1633 | - | 1e-92 | 68.92% (251) | transport protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2029 | - | 6e-93 | 69.72% (251) | hypothetical protein Mvan_2029 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4317|M.gilvum_PYR-GCK -----------------MLFAAGILGGLTGSIAGLASVATYPALLVAGLP
Mvan_2029|M.vanbaalenii_PYR-1 --------MPVSWSQAALLFAAGVIGGLTGSIAGLASVATYPALLLAGLP
MSMEG_2287|M.smegmatis_MC2_155 ----------MTVWDVVLLVFAGIAGGLTGSIAGLASVATYPALLVVGLP
TH_1633|M.thermoresistible__bu LRDLSRTGRHIRVTDVVLLLAAGIVGGLTGSIAGLASVATYPALLVVGLP
MLBr_00468|M.leprae_Br4923 ----------MLVSHLATIGLAGFGAGAINALVGSGTLITFPTLVALGYS
MAV_3519|M.avium_104 ---------------MFVICLAGLAAGAINALVGSGTLITFPTLVALGYP
MAB_3982|M.abscessus_ATCC_1997 ----------MPVQEMVLIALAAFGAGAINVVAGSGTLITFPTLIAFGYP
: *.. .* . :.* .:: *:*:*: * .
Mflv_4317|M.gilvum_PYR-GCK PVTANVTNTVALVFNGVGSVLGSRPELRGQRTLLTRAIPFAVLGGVTGAV
Mvan_2029|M.vanbaalenii_PYR-1 PVAANVTNTVALVANGVGSVWGSRPELAGQSALLKRTLPFAAVGGVTGAV
MSMEG_2287|M.smegmatis_MC2_155 PVAANVTNTVAVVFNGVGSIAGSRPELAGQGAWLKRIIPVAALGGVAGAA
TH_1633|M.thermoresistible__bu PVTANVTNTVALVFNGVGSVWGSRPELAGRRAWIMRLVPGAALGGLAGAV
MLBr_00468|M.leprae_Br4923 PVTSTMSNAVGLVAGGVSATWGYRAELGDRWSRLRWQIPMSLTGAALGAF
MAV_3519|M.avium_104 PVTATMSNATGLVAGSVSGTWGYRAELRGQWDRLRWQLPASLAGAGLGAY
MAB_3982|M.abscessus_ATCC_1997 PVVATMSNAVGLTVGSMTGVWGYRRELSGQWSRLKWQIPAVLLGALIGSF
**.:.::*:..:. ..: . * * ** .: : :* *. *:
Mflv_4317|M.gilvum_PYR-GCK LLLSTPAEGFEKIVPALLGFASLAILLP-------VRPAGDPSTASRRRR
Mvan_2029|M.vanbaalenii_PYR-1 LLLSTPAEGFEKLVPVLLGFASLAILLP-------VRPRRD-GTSSPRRR
MSMEG_2287|M.smegmatis_MC2_155 LLLSTPAEGFEKIVPFLLGFASVAILLP-------RREHRSARVANHRDH
TH_1633|M.thermoresistible__bu LLLSIPPEGFENSVPPLLALAALAIAVP-------RRADRDQTGAGHSRL
MLBr_00468|M.leprae_Br4923 LLLHLPEKVFTAIVPMLLVLALVLVVAGPRIQSWTRCRAKAAVNRAEHTT
MAV_3519|M.avium_104 LLLHLPEKVFAEIVPVLLVAALVLVVTGPRIQAWTARRAELAGRSPEHVP
MAB_3982|M.abscessus_ATCC_1997 LLLHLPEKTFVTVVPVLLILALVLVVVGPKIQAWARRRSEAQGRDADHVP
*** * : * ** ** * : :
Mflv_4317|M.gilvum_PYR-GCK TLTLAVQGLAVFAICIYGGYFGAAAGVLLLAMFLRLGGETLAHANAAKNV
Mvan_2029|M.vanbaalenii_PYR-1 TVTLVLQGAAVFAICIYGGYFGAAAGVLLLALFLRLGGETLAHANAAKNV
MSMEG_2287|M.smegmatis_MC2_155 LIRTGVEAAAIFLITIYGGYFGAAAGVLLLALMLRAGGATLPHANAGKNV
TH_1633|M.thermoresistible__bu LM---LQAAAVFVVSIYSGYFGAAAGVLLLAVLLNLSDATLATANAGKNL
MLBr_00468|M.leprae_Br4923 PAQMAMLALGTFAVSVYGGYFTAGQGILLVSLMSILLPESMQRINAAKNL
MAV_3519|M.avium_104 PGRMATLVFGTFAVGVYGGYFTAAQGILLVGLMGALLPESVQRMNAAKNL
MAB_3982|M.abscessus_ATCC_1997 PGRMAALTGGNFLVGIYGGYFTAAQGIMQIAVMGALLPESMQRMNAAKNV
. * : :*.*** *. *:: :.:: :: **.**:
Mflv_4317|M.gilvum_PYR-GCK VLGVANGVAAVIFAVVAP--VQWVAVLALGAGCLVGSRIGPVVVRRTPDR
Mvan_2029|M.vanbaalenii_PYR-1 VLGAANGVAALIFAVVAP--VAWPAVLALGTGCLLGSRLGPVVVRHAPDQ
MSMEG_2287|M.smegmatis_MC2_155 ILGVANLVASAIFVVFAP--VYWPAVVPLGIGCLIGSRLGPIIVRHAPST
TH_1633|M.thermoresistible__bu TLGVANTVAAAVFAVVAP--VQWAAVAPLGAGCLIGSRLGPVVVRHAPAT
MLBr_00468|M.leprae_Br4923 LSMLVNLTAAVGYTLVAFDRINWSAAGMIAASSLVGGLVGTRYARRLSPN
MAV_3519|M.avium_104 LALVVNVVAAVAYTSVAFGRISWAAAGLIAAGSLVGGLLGARYGRRLSGG
MAB_3982|M.abscessus_ATCC_1997 LAMITNIVAALAYTIVAFDRVSWTAAGIIAAGSFVGGLAGARWGRRLSPG
.* .*: :. .* : * *. :. ..::*. *. *: .
Mflv_4317|M.gilvum_PYR-GCK PMRLAIGLAGVALAVKLGIDAYR
Mvan_2029|M.vanbaalenii_PYR-1 PLRVAIGVAGVALAVKLGFDAYG
MSMEG_2287|M.smegmatis_MC2_155 PLRWLIGVAGIALAIKLALDTY-
TH_1633|M.thermoresistible__bu PLRLLIAASGLALAVKLGLDTYL
MLBr_00468|M.leprae_Br4923 VLRATIVAIGLIGLVRLLAV---
MAV_3519|M.avium_104 ALRAIIVVVGLIGLYRLLAVA--
MAB_3982|M.abscessus_ATCC_1997 ALRAVIVAVGLIGLWRLLF----
:* * *: :*