For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LDPAVWGAGTILGADLPRQINHGVDDVAVNLLRYLGHGATLVSGPAGQPVLLAFAERRLFAVLVLTIRDG RILKIEASVDPSAAERRRSGPVEF*
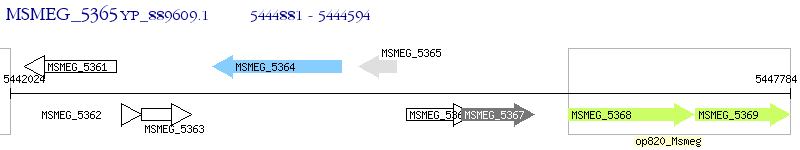
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5365 | - | - | 100% (95) | RNA polymerase sigma-70 factor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1221 | sigI | 3e-08 | 44.93% (69) | RNA polymerase sigma factor SigI |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv1189 | sigI | 3e-08 | 44.93% (69) | RNA polymerase sigma factor SigI |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2999c | sigI | 1e-27 | 67.05% (88) | RNA polymerase sigma factor SigI |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_0164 | sigI | 3e-26 | 65.88% (85) | RNA polymerase sigma factor SigI |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5365|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2999c|M.abscessus_ATCC_199 MSSSSTHDALVTAAWREHYPYLVNLAYQMLGDIGDAEDAVQEAFVRLARA
MAV_0164|M.avium_104 MNRSQSADDQVAEAWRRHRPYLVNLAYQMLGDIGDAEDVAQEAFLRLSAA
Mb1221|M.bovis_AF2122/97 ----MSQHDPVSAAWRAHRAYLVDLAFRMVGDIGVAEDMVQEAFSRLLRA
Rv1189|M.tuberculosis_H37Rv ----MSQHDPVSAAWRAHRAYLVDLAFRMVGDIGVAEDMVQEAFSRLLRA
MSMEG_5365|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2999c|M.abscessus_ATCC_199 EETEIDEPRAWLSVVVGRLCLDQLRSARMRRERTGVASVVESAAPLHVPA
MAV_0164|M.avium_104 G--DIDDVRGWLTVVASRLCLDQLRSARARHERPGDVGEQP------APP
Mb1221|M.bovis_AF2122/97 PVGDIDDERGWLIVVTSRLCLDHIKSASTRRERPQDIAAWHDG--DASVS
Rv1189|M.tuberculosis_H37Rv PVGDIDDERGWLIVVTSRLCLDHIKSASTRRERPQDIAAWHDG--DASVS
MSMEG_5365|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2999c|M.abscessus_ATCC_199 VTDPADRVTLDDEVRGALLEVLRSLSPGERVAFVLHDVFAVPFEEIASTV
MAV_0164|M.avium_104 RFDPADRITLDDEVRTALLEVLRRLSPGERVSFVLHDVFGVPFEAIAQTV
Mb1221|M.bovis_AF2122/97 SVDPADRVTLDDEVRLALLIMLERLGPAERVVFVLHEIFGLPYQQIATTI
Rv1189|M.tuberculosis_H37Rv SVDPADRVTLDDEVRLALLIMLERLGPAERVVFVLHEIFGLPYQQIATTI
MSMEG_5365|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2999c|M.abscessus_ATCC_199 GRPVGTCRQLARRARTRFQQSAARSTDVTGAEHQQLIEKFVTACAQGDLT
MAV_0164|M.avium_104 GRPVGTCRQLARRARAKFVAAQPKLNTVGAAEHQRVTEKFITACANGDLA
Mb1221|M.bovis_AF2122/97 GSQASTCRQLAHRARRKINESR-IAASVEPAQHRVVTRAFIEACSNGDLD
Rv1189|M.tuberculosis_H37Rv GSQASTCRQLAHRARRKINESR-IAASVEPAQHRVVTRAFIEACSNGDLD
MSMEG_5365|M.smegmatis_MC2_155 ----MLDPAVWGAGTILGADLPR-QINHGVDDVAVNLLRYLGHGATLVSG
MAB_2999c|M.abscessus_ATCC_199 GLMAVLDPGVWGVGTILGDPAPPPQVNRGRTDVATNLLVYLGPGATLVGG
MAV_0164|M.avium_104 GLAAVLDPTVWGVGTVLADPAPPPQVNHGPDAVATNLLRYLWPDVTLVGG
Mb1221|M.bovis_AF2122/97 TLLEVLDPGVAG-----EIDARKGVVVVGADRVGPTILRHWSHPATVLVA
Rv1189|M.tuberculosis_H37Rv TLLEVLDPGVAG-----EIDARKGVVVVGADRVGPTILRHWSHPATVLVA
:*** * * : * *. .:* : .*:: .
MSMEG_5365|M.smegmatis_MC2_155 P--AGQPVLLAFAERRLFAVLVLTIRDGRILKIEASVDPSAAERRRSGPV
MAB_2999c|M.abscessus_ATCC_199 P--FGQPVLLAFTQRRLFAVVTLTVREGLVLKIEATADPAARGR------
MAV_0164|M.avium_104 P--AGAPVLLAFSRRRLFAVIVLSIRDARVVKIEAIADPAARSAG-----
Mb1221|M.bovis_AF2122/97 QPVCGQPAVLAFVNRALAGVLALSIEAGKITKIHVLVQPSTLDPLRAELG
Rv1189|M.tuberculosis_H37Rv QPVCGQPAVLAFVNRALAGVLALSIEAGKITKIHVLVQPSTLDPLRAELG
* *.:*** .* * .*:.*::. . : **.. .:*::
MSMEG_5365|M.smegmatis_MC2_155 EF
MAB_2999c|M.abscessus_ATCC_199 --
MAV_0164|M.avium_104 --
Mb1221|M.bovis_AF2122/97 GG
Rv1189|M.tuberculosis_H37Rv GG