For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DLKKLAVTTTMTGALGLGALGLGMGAAHADPATPDPRPAPVVPDVNVPGVNVPDVNVPGVNVPDVNVPGV NVPDVNVPGVNVPDVNVPAVNVPDVNVPAVNVPDVNVPGVNLPDVNLPEVNFPEVNGYFRLPDTQIPPGQ LMTERIINGVENPFFGIPPGQLKKLPTVNGLVNPFFDERPGHWVIPGDPA*
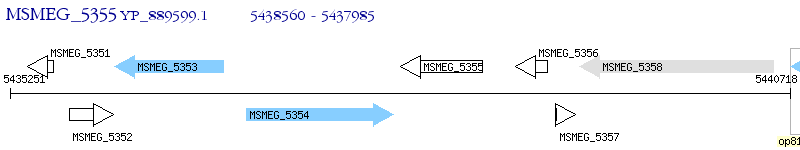
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5355 | - | - | 100% (191) | hypothetical protein MSMEG_5355 |
| M. smegmatis MC2 155 | MSMEG_6405 | - | 3e-08 | 27.72% (101) | Erp protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1782c | PPE24 | 1e-12 | 25.90% (139) | PPE family protein |
| M. gilvum PYR-GCK | Mflv_1158 | - | 1e-08 | 27.43% (113) | repeat of unknown function XGLTT |
| M. tuberculosis H37Rv | Rv1753c | PPE24 | 1e-12 | 25.90% (139) | PPE family protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2580c | - | 1e-06 | 39.19% (74) | hypothetical protein MAB_2580c |
| M. marinum M | MMAR_1028 | - | 5e-14 | 28.07% (171) | PPE family protein |
| M. avium 104 | MAV_0207 | - | 1e-07 | 25.66% (113) | Erp protein |
| M. thermoresistible (build 8) | TH_2084 | - | 5e-10 | 24.86% (173) | Erp protein |
| M. ulcerans Agy99 | MUL_1434 | - | 4e-06 | 32.56% (86) | hypothetical protein MUL_1434 |
| M. vanbaalenii PYR-1 | Mvan_5651 | - | 5e-08 | 26.42% (106) | exported repetitive protein precursor PirG (cell surface |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 MNFSVLPPEINSALIFAGAGPEPMAAAATAWDGLAMELASAAASFGSVTS
Rv1753c|M.tuberculosis_H37Rv MNFSVLPPEINSALIFAGAGPEPMAAAATAWDGLAMELASAAASFGSVTS
MMAR_1028|M.marinum_M MNFSVLPPEVNSALIFTGPGAEPMRTAATAWGGMAVELESAADSFASVTS
Mflv_1158|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5651|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2084|M.thermoresistible__bu --------------------------------------------------
MAV_0207|M.avium_104 --------------------------------------------------
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 GLVGGAWQGASSSAMAAAAAPYAAWLAAAAVQAEQTAAQAAAMIAEFEAV
Rv1753c|M.tuberculosis_H37Rv GLVGGAWQGASSSAMAAAAAPYAAWLAAAAVQAEQTAAQAAAMIAEFEAV
MMAR_1028|M.marinum_M GLVGGAWQGASSVAMGTAAASYLGWLNAAAAHAEQAAAQAEALVAEFAAA
Mflv_1158|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5651|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2084|M.thermoresistible__bu --------------------------------------------------
MAV_0207|M.avium_104 --------------------------------------------------
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 KTAVVQPMLVAANRADLVSLVMSNLFGQNAPAIAAIEATYEQMWAADVSA
Rv1753c|M.tuberculosis_H37Rv KTAVVQPMLVAANRADLVSLVMSNLFGQNAPAIAAIEATYEQMWAADVSA
MMAR_1028|M.marinum_M QSAMVQPFLLAANRSSLVSLVLSNLFGQNAPAIAAVEAEYEQMWALDVST
Mflv_1158|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5651|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2084|M.thermoresistible__bu --------------------------------------------------
MAV_0207|M.avium_104 --------------------------------------------------
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 MSAYHAGASAIASALSPFSKPLQNLAGLP----AWLASGAPAAAMTAAAG
Rv1753c|M.tuberculosis_H37Rv MSAYHAGASAIASALSPFSKPLQNLAGLP----AWLASGAPAAAMTAAAG
MMAR_1028|M.marinum_M MATYHAGASAAAASLTPLAQPLQNLSDLPGRVIAAFGNGAPATTVTAAVT
Mflv_1158|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5651|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2084|M.thermoresistible__bu --------------------------------------------------
MAV_0207|M.avium_104 --------------------------------------------------
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 IPALAGGPTAINLGIANVGGGNVGNANNGLANIGNANLGNYNFGSGNFGN
Rv1753c|M.tuberculosis_H37Rv IPALAGGPTAINLGIANVGGGNVGNANNGLANIGNANLGNYNFGSGNFGN
MMAR_1028|M.marinum_M ALRLPADASTVNFGLANIGSGNVGNANNGIGNLGSGNLGNFNLGSGNLGS
Mflv_1158|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5651|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2084|M.thermoresistible__bu --------------------------------------------------
MAV_0207|M.avium_104 --------------------------------------------------
MSMEG_5355|M.smegmatis_MC2_155 ---------------------------------------------MDLKK
MUL_1434|M.ulcerans_Agy99 ---------------------------------------------MQLRR
MAB_2580c|M.abscessus_ATCC_199 ---------------------------------------------MKMAR
Mb1782c|M.bovis_AF2122/97 SNIGSASLGNNNIGFGNLGSNNVGVGNLGNLNTGFANTGLGNFGFGNTGN
Rv1753c|M.tuberculosis_H37Rv SNIGSASLGNNNIGFGNLGSNNVGVGNLGNLNTGFANTGLGNFGFGNTGN
MMAR_1028|M.marinum_M ANAGWANLGSNNIGGANSGGNNLGWGNLGGLNTGFANAGSGNFGFANTGN
Mflv_1158|M.gilvum_PYR-GCK --------------------------------------------MPNRRR
Mvan_5651|M.vanbaalenii_PYR-1 --------------------------------------------MPNRRR
TH_2084|M.thermoresistible__bu --------------------------------------------------
MAV_0207|M.avium_104 --------------------------------------------MPNRRR
MSMEG_5355|M.smegmatis_MC2_155 LAVTTTMTGALGLGALGLGMGAAHADPATP--------------------
MUL_1434|M.ulcerans_Agy99 IYLCT----ILAAAAAGVAATLAAFPASSP--------------------
MAB_2580c|M.abscessus_ATCC_199 ALTYGAVAGGLTLGILGFGASVASAEPPPP--------------------
Mb1782c|M.bovis_AF2122/97 NNIGIGLTGNNQIGIGGLNSGTGNFGLFNSGSGNVGFFNSGNGNFGIGNS
Rv1753c|M.tuberculosis_H37Rv NNIGIGLTGNNQIGIGGLNSGTGNFGLFNSGSGNVGFFNSGNGNFGIGNS
MMAR_1028|M.marinum_M NNIGIGLTGDNQVGIGGLNSGVGNFGLFNSGTGNVGLFNSGTGNFGIGNS
Mflv_1158|M.gilvum_PYR-GCK RRLSTALSTVAALAVASPIAVIAVSELSAST-------------------
Mvan_5651|M.vanbaalenii_PYR-1 RRLSTALSTVAALAVASPVAVVAITELSAE--------------------
TH_2084|M.thermoresistible__bu ------MSTVAALAVASPFAAIAVTELVSS--------------------
MAV_0207|M.avium_104 RKLSTAMSAVAALAVASPCAYFLVYESTAG--------------------
. .
MSMEG_5355|M.smegmatis_MC2_155 --------DPRPAPVVPDVNVPGVNVPDVNVPGVNVPDVNVPGVNVP---
MUL_1434|M.ulcerans_Agy99 --------HIQPS-----------SAPIHDLP---LDDLGIPGVSV----
MAB_2580c|M.abscessus_ATCC_199 --------IPVPG------------LPGPGGP----PGPGIP--------
Mb1782c|M.bovis_AF2122/97 GNFNTGGWNSGHG------NTGFFNAGSFNTGMLDVGNANTGSLNTGSYN
Rv1753c|M.tuberculosis_H37Rv GNFNTGGWNSGHG------NTGFFNAGSFNTGMLDVGNANTGSLNTGSYN
MMAR_1028|M.marinum_M GAYGTGLFNSGQA------NTGLLNAGSFNTGLLDVGNANTGSLNTGSFN
Mflv_1158|M.gilvum_PYR-GCK ----LAGRDGAPQ------HREFVQAATIT----DLPGELMTALSQGLAQ
Mvan_5651|M.vanbaalenii_PYR-1 -----PGRDGAPQ------HREFVQAATIT----DLPGELFNALSQGLSQ
TH_2084|M.thermoresistible__bu -------TQPAPQ------QREFVQASLLT----DLPTELMGALSEGLAQ
MAV_0207|M.avium_104 --------NKAPE------HHEFKQAAVMS----DLPGELMGALSQGLSQ
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 MGDFNPGSSNTGTFNTGNANTGFLNAGNINTGVFNIGHMNNGLFNTGDMN
Rv1753c|M.tuberculosis_H37Rv MGDFNPGSSNTGTFNTGNANTGFLNAGNINTGVFNIGHMNNGLFNTGDMN
MMAR_1028|M.marinum_M MGSFNPGPSNTGALNTGTANTGWFNTGSLNTGAFNIGDMNNGLFNTGEMN
Mflv_1158|M.gilvum_PYR-GCK FG-------------------------------INLPPLPTGLLTGTGA-
Mvan_5651|M.vanbaalenii_PYR-1 FG-------------------------------INLPPMPTGLLTGSGA-
TH_2084|M.thermoresistible__bu FG-------------------------------INLPPMPS--LTGSGAS
MAV_0207|M.avium_104 FG-------------------------------INLPPVPA--LSGGAT-
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 NGVFYRGVGQGSLQFSITTPDLTLPPLQIPGISVP---------------
Rv1753c|M.tuberculosis_H37Rv NGVFYRGVGQGSLQFSITTPDLTLPPLQIPGISVP---------------
MMAR_1028|M.marinum_M NGIFYRGVGQGNFYFSITTPDLTLPPLEIPGITIPNLSLPPITVPALTIP
Mflv_1158|M.gilvum_PYR-GCK -----------------AAPTTLSPGLGTAGLTSP---------------
Mvan_5651|M.vanbaalenii_PYR-1 -----------------TSPTTLSPGLGFPSLASP---------------
TH_2084|M.thermoresistible__bu M------------SPGLTSPGLTSPGLTSPGLTSP---------------
MAV_0207|M.avium_104 -----------------STPGLASPGLGSPGLGSP---------------
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 --------------------------------------------------
Rv1753c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1028|M.marinum_M AATTPANVVVGGFDLPGVTLPALTIPAATTPANVVVGGFDLPGVTLPALT
Mflv_1158|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5651|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2084|M.thermoresistible__bu --------------------------------------------------
MAV_0207|M.avium_104 --------------------------------------------------
MSMEG_5355|M.smegmatis_MC2_155 ------------------DVNVPGVNVPDVNVPA------VNVPDVNVPA
MUL_1434|M.ulcerans_Agy99 ------------------DIDAK---VPNLNSVE------QVVPNVSVPN
MAB_2580c|M.abscessus_ATCC_199 --------------------------GPGVPGPG------PGIPGPGVPG
Mb1782c|M.bovis_AF2122/97 -------------AFSLPAITLPSLTIPAATTPANITVGAFSLPGLTLPS
Rv1753c|M.tuberculosis_H37Rv -------------AFSLPAITLPSLNIPAATTPANITVGAFSLPGLTLPS
MMAR_1028|M.marinum_M IPAATTPANVVVGGFDLPGVTLPALTIPAATTPANVVVGGFDLPGVTLPA
Mflv_1158|M.gilvum_PYR-GCK ------------------GLMPGLSAAPSLTDPS------LAN-----PA
Mvan_5651|M.vanbaalenii_PYR-1 ------------------GLTTPGLTAPTLTDPG------LANPALTNPA
TH_2084|M.thermoresistible__bu ------------------GLTSPGLTSPGLTSPG------LTNPGLTSPG
MAV_0207|M.avium_104 ------------------GLGTPGLGTPGLTNPG------LTSPGATSPG
* *
MSMEG_5355|M.smegmatis_MC2_155 VNVPDVNVPG------VNLPDVNLPE--VNFPEVNGYFR-----------
MUL_1434|M.ulcerans_Agy99 INLPNVKVN-----------------------------------------
MAB_2580c|M.abscessus_ATCC_199 PDGPGPDGP-----------------------------------------
Mb1782c|M.bovis_AF2122/97 LNIPAATTPANITVGAFSLPGLTLPS--LNIPAATTPANITVGAFSLP--
Rv1753c|M.tuberculosis_H37Rv LNIPAATTPANITVGAFSLPGLTLPS--LNIPAATTPANITVGAFSLPGL
MMAR_1028|M.marinum_M LTIPAATTPANVVVGGFDLPGVTLPA--LTIPAATTPANVVVGGFDLPGV
Mflv_1158|M.gilvum_PYR-GCK LTSPIGATPG------LTTPGLTTPG--LTTPGLT---------------
Mvan_5651|M.vanbaalenii_PYR-1 LTSPTGAAPG------LTTPGLTTPG--LTTPGLT---------------
TH_2084|M.thermoresistible__bu LTDPALTTPG------LTTPGLTTPG--LTTPGLTTPG------------
MAV_0207|M.avium_104 LTSPGLTSPG------LTSPGLTSPGAAPTTPGLTAPG------------
*
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 --------------------------------------------------
Rv1753c|M.tuberculosis_H37Rv TLPSLNIPAATTPANITVGAFSLPGLTLPSLNIPAATTPANITVGAFSLP
MMAR_1028|M.marinum_M TLPALTIPAATTPANVVVGGFDLPGVTLPALTIPAATTPANVVVGGFDLP
Mflv_1158|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5651|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2084|M.thermoresistible__bu --------------------------------------------------
MAV_0207|M.avium_104 --------------------------------------------------
MSMEG_5355|M.smegmatis_MC2_155 --LPDTQIPPGQLMTERI--------------------------------
MUL_1434|M.ulcerans_Agy99 ---------PG---------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 GLTLPSLNIPAATTPANITVSGFQLPPLSIPSVAIPPVTVPP--------
Rv1753c|M.tuberculosis_H37Rv GLTLPSLNIPAATTPANITVSGFQLPPLSIPSVAIPPVTVPP--------
MMAR_1028|M.marinum_M GVTLPALTIPAATTPANVVVGGFDLPQLGIPSLTIPPLTLAEGTTVGAFS
Mflv_1158|M.gilvum_PYR-GCK ---DPALASPGLTTP-----------------------------------
Mvan_5651|M.vanbaalenii_PYR-1 ---DPALASPGLTTP-----------------------------------
TH_2084|M.thermoresistible__bu --LDPAQTTPGLTTP-----------------------------------
MAV_0207|M.avium_104 --ALPTTPGGGVATPG----------------------------------
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 ---------------------------------------------ITVGA
Rv1753c|M.tuberculosis_H37Rv ---------------------------------------------ITVGA
MMAR_1028|M.marinum_M LPELGIPSLTIPPLTLAEGTTVGAFSLPELGIPSLTIPPLTLAEGTTVGA
Mflv_1158|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5651|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2084|M.thermoresistible__bu --------------------------------------------------
MAV_0207|M.avium_104 --------------------------------------------------
MSMEG_5355|M.smegmatis_MC2_155 --INGVENPFFGIPPGQLKKLPTVNGLVNPFFDERPGHWVIPGDPA----
MUL_1434|M.ulcerans_Agy99 ---------HVGLP-GRGR-------------------------------
MAB_2580c|M.abscessus_ATCC_199 ---------FFH--------------------------------------
Mb1782c|M.bovis_AF2122/97 FNLPPLQIPEVTIPQLTIPAGITIGGFSLPAIHTQPITVGQIGVGQFGLP
Rv1753c|M.tuberculosis_H37Rv FNLPPLQIPEVTIPQLTIPAGITIGGFSLPAIHTQPITVGQIGVGQFGLP
MMAR_1028|M.marinum_M FSLPQLGIPSLTIPSLTVPAGTTVGGFDLPALQTRSITVGGIDAGGFRTP
Mflv_1158|M.gilvum_PYR-GCK ----ALTDPGLTTPGLTTPGLTTP-GLTTPGLTTPAALT--DPALGLPAT
Mvan_5651|M.vanbaalenii_PYR-1 ----SLTDPALASPGLTTPGLSTPPALTDPALG-ALPIS--TSGLG---T
TH_2084|M.thermoresistible__bu ----GVSTPDLTNPALTSPLGEAPGGLTTPSVGLDPALTPISNTAGLPAP
MAV_0207|M.avium_104 ----AGLNPALSNPGLTSPAGTAP-GLGSPTVA----------------P
.
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 SIG-----------------------------------------------
Rv1753c|M.tuberculosis_H37Rv SIG-----------------------------------------------
MMAR_1028|M.marinum_M AISTVSSTGYLELPTVTIPGVRVNNFVAFIPNNIEFLQTGQPGVIPQIGG
Mflv_1158|M.gilvum_PYR-GCK GEIP----------------------------------------------
Mvan_5651|M.vanbaalenii_PYR-1 GEVP----------------------------------------------
TH_2084|M.thermoresistible__bu GEVP----------------------------------------------
MAV_0207|M.avium_104 SEVP----------------------------------------------
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 --WDVFLST---------------------------PRITVPAFGIP---
Rv1753c|M.tuberculosis_H37Rv --WDVFLST---------------------------PRITVPAFGIP---
MMAR_1028|M.marinum_M LEHQAYISTGDIVIQGFSISGIGFSLPQIDVGAFTLPELTIPPIGIPPTI
Mflv_1158|M.gilvum_PYR-GCK ---------------------------------------ISAPIGLDP--
Mvan_5651|M.vanbaalenii_PYR-1 ---------------------------------------ISAPIGLDP--
TH_2084|M.thermoresistible__bu ---------------------------------------ITAPLGPDP--
MAV_0207|M.avium_104 ---------------------------------------IDSGAGLDP--
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 ---FTLQFQTN----VPALQPPGGGLSTFTNGALIFGEFDLPQLVVHPYT
Rv1753c|M.tuberculosis_H37Rv ---FTLQFQTN----VPALQPPGGGLSTFTNGALIFGEFDLPQLVVHPYT
MMAR_1028|M.marinum_M VGAFTLPEITTPVITTPPLVVDPVSVAGFTLPEITTPEFTTPPLVVDPVS
Mflv_1158|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5651|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2084|M.thermoresistible__bu --------------------------------------------------
MAV_0207|M.avium_104 --------------------------------------------------
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 LTG-------------------PIVIGSFFLPAFNIPGIDVP--------
Rv1753c|M.tuberculosis_H37Rv LTG-------------------PIVIGSFFLPAFNIPGIDVP--------
MMAR_1028|M.marinum_M VAGFTLPEITTPEFTTPPLVVDPVSVAGFTLPEITTPEFTTPPLVVDPVS
Mflv_1158|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5651|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2084|M.thermoresistible__bu --------------------------------------------------
MAV_0207|M.avium_104 --------------------------------------------------
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 -----------------------------------------------AIN
Rv1753c|M.tuberculosis_H37Rv -----------------------------------------------AIN
MMAR_1028|M.marinum_M VAGFTLPQISTPEFTTAPFTIDPVSVAGFTLPQISTPEFTTAPFTIDPVS
Mflv_1158|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5651|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2084|M.thermoresistible__bu --------------------------------------------------
MAV_0207|M.avium_104 --------------------------------------------------
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 VDGFTLPQITTPAITTPEFAIPPIGVGGFTLPQITTQEIITPELTINSIG
Rv1753c|M.tuberculosis_H37Rv VDGFTLPQITTPAITTPEFAIPPIGVGGFTLPQITTQEIITPELTINSIG
MMAR_1028|M.marinum_M VAGFTLPQISTPEFTTAPFTIDPVSVAGFTLPQISTPEFTTAPFTIDPVS
Mflv_1158|M.gilvum_PYR-GCK ALG-SYPILGDPS-LAATQPVA---------TSSGG---LLGDLTSAANS
Mvan_5651|M.vanbaalenii_PYR-1 ALG-SYPVLGDPS-LAATQPVA---------SSSGG---LLGDLSSAANS
TH_2084|M.thermoresistible__bu TLG-TYPILGDPS-LAMAPPVA---------TSTGGGG-LISELSTAANQ
MAV_0207|M.avium_104 GAGGTYPILGDPSTFGNASPIGGGGTGLGGGSSSGGSGGLVNDVMQAANQ
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 VGGFTLPQITTPPITTPPLTIDPINLTGFTLPQITTPPITTPPLTIDPIN
Rv1753c|M.tuberculosis_H37Rv VG------------------------------------------------
MMAR_1028|M.marinum_M VAGFTLPQISTPEFTTAPFTIDPVSVAGFTLPQISTPEFTTAPFTIDPVS
Mflv_1158|M.gilvum_PYR-GCK LG------------------------------------------------
Mvan_5651|M.vanbaalenii_PYR-1 LG------------------------------------------------
TH_2084|M.thermoresistible__bu LG------------------------------------------------
MAV_0207|M.avium_104 LG------------------------------------------------
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 LTGFTLPQITTPPITTPPLTIDPINLTGFTLPQITTPPITTPPLTIDPIN
Rv1753c|M.tuberculosis_H37Rv --GFTLPQITTPPITTPPLTIDPINLTGFTLPQITTPPITTPPLTIDPIN
MMAR_1028|M.marinum_M VAGFTLPQISTPEFTTAPFTIDPVSVAGFTLPQISTPEFTTAPFTIDPVS
Mflv_1158|M.gilvum_PYR-GCK ----------------AGEAID---------------------------L
Mvan_5651|M.vanbaalenii_PYR-1 ----------------AGEAID---------------------------L
TH_2084|M.thermoresistible__bu ----------------AGQAID---------------------------L
MAV_0207|M.avium_104 ----------------AGQAID---------------------------L
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 LTGFTLPQITTPPITTPPLTIEPIGVGGFTTPPLTVPGIHLPSTTIGAFA
Rv1753c|M.tuberculosis_H37Rv LTGFTLPQITTPPITTPPLTIEPIGVGGFTTPPLTVPGIHLPSTTIGAFA
MMAR_1028|M.marinum_M VAGFTLPQISTPQFTTAPITIPPIGLGSFTTPPLTIPSIHLPGTTIGEFA
Mflv_1158|M.gilvum_PYR-GCK LKGVLMPAIMSAVKPPVPP----VPAAPLATAAGEV-APVEAGLGAAEAS
Mvan_5651|M.vanbaalenii_PYR-1 LKGVLMPAIMSAVKPPAP-----LPAAPVAQAAGTVPAEAEAALGAAEAA
TH_2084|M.thermoresistible__bu LKGVVMPAVTSAIKSAAQ------PPGP------DVAPPPAPEVPAPDA-
MAV_0207|M.avium_104 LKGLVMPAITQGMHGGAAAGALPGAAGALPGAAGALPGAAGALPGAAGAL
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 IPGGPGYFNSSTAPSSGFFNSGAGGNSGFGNNGSGLSGWFNTNPAGLLGG
Rv1753c|M.tuberculosis_H37Rv IPGGPGYFNSSTAPSSGFFNSGAGGNSGFGNNGSGLSGWFNTNPAGLLGG
MMAR_1028|M.marinum_M FPAGPGYLNSSATPSSGFFNSGAGGNSGLGNNGSGLSGWFNTNPVGLLGG
Mflv_1158|M.gilvum_PYR-GCK FETGAEPGA-----------------------------------------
Mvan_5651|M.vanbaalenii_PYR-1 IEPGA---------------------------------------------
TH_2084|M.thermoresistible__bu --------------------------------------------------
MAV_0207|M.avium_104 PAAAGAAGALPAAAGAAPALPPV---------------------------
MSMEG_5355|M.smegmatis_MC2_155 --------------------------------------------------
MUL_1434|M.ulcerans_Agy99 --------------------------------------------------
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 SGYQNFGGLSSGFSNLGSGVSGFANRGILPFSVASVVSGFANIGTNLAGF
Rv1753c|M.tuberculosis_H37Rv SGYQNFGGLSSGFSNLGSGVSGFANRGILPFSVASVVSGFANIGTNLAGF
MMAR_1028|M.marinum_M SGYQNYGGLSSGFSNLGSGVSGFANTGVRPFSLTGLISGFANIGSNLSGF
Mflv_1158|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5651|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2084|M.thermoresistible__bu --------------------------------------------------
MAV_0207|M.avium_104 --------------------------------------------------
MSMEG_5355|M.smegmatis_MC2_155 ------
MUL_1434|M.ulcerans_Agy99 ------
MAB_2580c|M.abscessus_ATCC_199 ------
Mb1782c|M.bovis_AF2122/97 FQGTTS
Rv1753c|M.tuberculosis_H37Rv FQGTTS
MMAR_1028|M.marinum_M FGSNVP
Mflv_1158|M.gilvum_PYR-GCK ------
Mvan_5651|M.vanbaalenii_PYR-1 ------
TH_2084|M.thermoresistible__bu ------
MAV_0207|M.avium_104 ------