For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RHSHVRPIGLTNGGLMSINAVRTGHEPASYSVRLSSEVVEDALRLRHEALAGHGLPAPALDGDRFDEHCL HVLVRDDASGELVGSCRILPHDGATDAGGLYLATAFDVSALDPLRPAALEFSRAVIRPGHRNSRAALALW SGVLGYAEQHGYAHLVGSVPMPLHPAGEVRAMRNLLRREHAAPGRYTVRPYRPVVHDALHLPGHEPQCTL PEPLAAQVRLGARLCGDPAYDPEFGVAHFPMLQFLRPS*
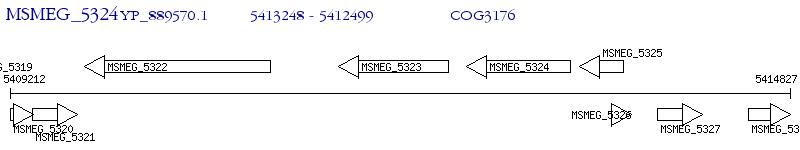
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5324 | - | - | 100% (249) | hypothetical protein MSMEG_5324 |
| M. smegmatis MC2 155 | MSMEG_5323 | - | 6e-53 | 44.94% (247) | hypothetical protein MSMEG_5323 |
| M. smegmatis MC2 155 | MSMEG_2355 | - | 4e-51 | 47.51% (221) | hypothetical protein MSMEG_2355 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3053c | - | 3e-49 | 46.79% (218) | hypothetical protein Mb3053c |
| M. gilvum PYR-GCK | Mflv_4257 | - | 2e-53 | 45.78% (249) | hypothetical protein Mflv_4257 |
| M. tuberculosis H37Rv | Rv3027c | - | 3e-49 | 46.79% (218) | hypothetical protein Rv3027c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3359c | - | 4e-46 | 46.84% (190) | hypothetical protein MAB_3359c |
| M. marinum M | MMAR_1686 | - | 6e-47 | 45.02% (231) | hypothetical protein MMAR_1686 |
| M. avium 104 | MAV_3874 | - | 5e-50 | 44.98% (229) | hypothetical protein MAV_3874 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1924 | - | 4e-47 | 45.02% (231) | hypothetical protein MUL_1924 |
| M. vanbaalenii PYR-1 | Mvan_2103 | - | 2e-55 | 47.76% (245) | hypothetical protein Mvan_2103 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3053c|M.bovis_AF2122/97 ----------------------------------------MVEAAQRLRY
Rv3027c|M.tuberculosis_H37Rv ----------------------------------------MVEAAQRLRY
MMAR_1686|M.marinum_M MSIASVLIPSDKPR----GPATDAGAGPR-YSLLLSTDSALIEAAQRLRY
MUL_1924|M.ulcerans_Agy99 MSIASVLIPSDKPR----GPATDAGAGPC-YSLLLSTDSALIEAAQRLRY
MAV_3874|M.avium_104 MSIASVLIPSDHPS----GPATGSSAAPR-YSLLLSADPAMIEAAQRLRY
Mflv_4257|M.gilvum_PYR-GCK MNASSVLIPSDHS--------VTAGSVPR-YSMLLCTDPASIEAAQRLRY
MAB_3359c|M.abscessus_ATCC_199 MSTASVLIAADQSMPEVAGKTGEAALGPR-YTLLLSNDSADIDAVQRLRY
Mvan_2103|M.vanbaalenii_PYR-1 MSTASVLIAPDDID---HAQGGSSPSAPR-YTLLLCTDVDSIEAAQRLRH
MSMEG_5324|M.smegmatis_MC2_155 MRHSHVRPIGLTNGGLMSINAVRTGHEPASYSVRLS--SEVVEDALRLRH
:: . ***:
Mb3053c|M.bovis_AF2122/97 DVFSTTPGFALP------AAADTRRDGDRFDEYCDHLLVRDDDTGELVGC
Rv3027c|M.tuberculosis_H37Rv DVFSTTPGFALP------AAADTRRDGDRFDEYCDHLLVRDDDTGELVGC
MMAR_1686|M.marinum_M DVFAATPGFALP------AADD-RRDVDRFDEYCDHLLVRDDDTGELVGC
MUL_1924|M.ulcerans_Agy99 DVFAATPGFALP------AADD-RRDVDRFDEYCDHLLVRDDDTGELVGC
MAV_3874|M.avium_104 EVFTSTPGFALP------SADGSGRDVDRFDEFCDHLLVRDDDTGELVGC
Mflv_4257|M.gilvum_PYR-GCK DVFTSEPGYTLNRGTLHRSAAPDGLDADRFDEFCDHLLVRDDDTGQLVGC
MAB_3359c|M.abscessus_ATCC_199 DVFSSEPGFQLAS----TPEAFEGRDADRFDEHCDHLLVREDVSGDVVGC
Mvan_2103|M.vanbaalenii_PYR-1 DVFSTEPGFTLTD-----DGGSGGLDADRFDEYCDHLLVRDDATGELVGC
MSMEG_5324|M.smegmatis_MC2_155 EALAG-HGLPAP-----------ALDGDRFDEHCLHVLVRDDASGELVGS
:.:: * * *****.* *:***:* :*::**.
Mb3053c|M.bovis_AF2122/97 YRMLAPAGAIAAGGLYTATEFDVCAFDPLRPSLVEMGRAVVREGHRNGGV
Rv3027c|M.tuberculosis_H37Rv YRMLAPAGAIAAGGLYTATEFDVCAFDPLRPSLVEMGRAVVREGHRNGGV
MMAR_1686|M.marinum_M YRMLPPAGAIAAGGLYTATEFDVGAFDPLRPSLVEMGRAAVRDGHRNGAV
MUL_1924|M.ulcerans_Agy99 YRMLPPAGAIAAGGLYTATEFDVGAFDPLRPSLVEMGRAAVRDGHRNGAV
MAV_3874|M.avium_104 YRMLAPAGAIAAGGLYTATEFDIRAFDPLRPSLVEMGRAVVRDGHRNGGV
Mflv_4257|M.gilvum_PYR-GCK YRMLPPPGAIAAGGLYTATEFDISALDALRPSLVEMGRAVVRDGHRNGAV
MAB_3359c|M.abscessus_ATCC_199 YRMLPPPGAIAAGGLYSATEFDISGLDSLRPQLVEMGRAVVRGDHRNGAV
Mvan_2103|M.vanbaalenii_PYR-1 YRMLPPPGAIAAGGLYTATEFDVTALDPLRPSLVEMGRAVVRDGHRNGAV
MSMEG_5324|M.smegmatis_MC2_155 CRILPHDGATDAGGLYLATAFDVSALDPLRPAALEFSRAVIRPGHRNSRA
*:*. ** ***** ** **: .:*.*** :*:.**.:* .***. .
Mb3053c|M.bovis_AF2122/97 VLLMWAGILAYLDRYGYDYVTGCVSVPI----GGDGETP-GSRLRGVRDF
Rv3027c|M.tuberculosis_H37Rv VLLMWAGILAYLDRYGYDYVTGCVSVPI----GGDGETP-GSRLRGVRDF
MMAR_1686|M.marinum_M VLLMWAGILAYLDRCDYDYVTGCVSVPIVEPTGRDGITP-GRELRGVRDF
MUL_1924|M.ulcerans_Agy99 VLLMWAGILAYLDRCDYDYVTGCVSVPIVEPTGRDGITP-GRELRGVRDF
MAV_3874|M.avium_104 VLLMWAGILAYLDRYGYDYVTGCVSVPIG---DADDAPP-GSRLRGVRDF
Mflv_4257|M.gilvum_PYR-GCK VLLMWTGILAYLDRSGYDYVTGCVSVPVA---DPDGLTPPGSRLRGVRDL
MAB_3359c|M.abscessus_ATCC_199 VLLMWAGILAYLDRCDYTYVTGCVSVPMQ---DSPEQAP-GSQVRAVRDF
Mvan_2103|M.vanbaalenii_PYR-1 VLLMWAGILAYLDHSGYDYVAGCVSVPVA---DPEGRSPAGSQLRGVRDF
MSMEG_5324|M.smegmatis_MC2_155 ALALWSGVLGYAEQHGYAHLVGSVPMPLH---------P-AGEVRAMRNL
.* :*:*:*.* :: .* ::.*.*.:*: * . .:*.:*::
Mb3053c|M.bovis_AF2122/97 ILNRHAAPPQCQVYPYRPVRVDGRSLDDILPPPRPAVPPLMRGYLRLGAR
Rv3027c|M.tuberculosis_H37Rv ILNRHAAPPQCQVYPYRPVRVDGRSLDDILPPPRPAVPPLMRGYLRLGAR
MMAR_1686|M.marinum_M VLRRHPAS--YRVDPYRPVRIDGKSLDDIAPPPRPAIPPLLRGYLRLGAQ
MUL_1924|M.ulcerans_Agy99 VLRRHPAS--YRVDPYRPVRIDGKSLDDIAPPPRPAIPPLLRGYLRLGAQ
MAV_3874|M.avium_104 VVSRHGAPARYRVRPHRPVVVDGTALDDIPPPARPSVPALMRGYLRLGAQ
Mflv_4257|M.gilvum_PYR-GCK VRRRNAAPPEYTVRPYRPVVIDGHHLDDIAAPPRVKVPALLRGYLRLGAQ
MAB_3359c|M.abscessus_ATCC_199 VMKRHAAAPQYTVRPYNPVVVGGMGLDDIAPPAKVQVPALMRGYLRLGAK
Mvan_2103|M.vanbaalenii_PYR-1 VIRRHAAPAEYTVRPYRPVVVDGRPLDEIEPPARLSVPALMRGYLRLGAQ
MSMEG_5324|M.smegmatis_MC2_155 LRREHAAPGRYTVRPYRPVVHDALHLPGHEP--QCTLPEPLAAQVRLGAR
: .: *. * *:.** .. * . : :* : . :****:
Mb3053c|M.bovis_AF2122/97 ACGEPAHDPDFGVGDFCLLLDKDHADTRYLRRLRSVAAASEMVNDAR---
Rv3027c|M.tuberculosis_H37Rv ACGEPAHDPDFGVGDFCLLLDKDHADTRYLRRLRSVAAASEMVNDAR---
MMAR_1686|M.marinum_M ACGEPAHDPDFGVGDFCVLLGKRDADIRYLKRLRSVSAAAEMAGELAGGA
MUL_1924|M.ulcerans_Agy99 ACGEPAHDPDFGVGDFCVLLGKRDADIRYLKRLRSVSAAAEMAGELAGGA
MAV_3874|M.avium_104 VCGEPAHDPDFGVGDFCVLLGKQDADTRYLKRLRSVSAAAELAGGR----
Mflv_4257|M.gilvum_PYR-GCK VCGEPAHDPDFGVGDFPALLDKRRADVRYLTRLRSAAAAAADVTR-----
MAB_3359c|M.abscessus_ATCC_199 VLGEPAHDPDFGVADFMALLNKNEADVRYLKRLRSVGAASEIGATTVPSE
Mvan_2103|M.vanbaalenii_PYR-1 VCGEPAHDPDFGVGDFPALLAKRHADTRYLRRLRSVGMATAAVADGTRS-
MSMEG_5324|M.smegmatis_MC2_155 LCGDPAYDPEFGVAHFPMLQFLRPS-------------------------
*:**:**:***..* * :
Mb3053c|M.bovis_AF2122/97 --
Rv3027c|M.tuberculosis_H37Rv --
MMAR_1686|M.marinum_M A-
MUL_1924|M.ulcerans_Agy99 A-
MAV_3874|M.avium_104 --
Mflv_4257|M.gilvum_PYR-GCK --
MAB_3359c|M.abscessus_ATCC_199 SE
Mvan_2103|M.vanbaalenii_PYR-1 --
MSMEG_5324|M.smegmatis_MC2_155 --