For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPEAVIVATARSPIGRAGKGSLVEMRPDDLAAQMVKAVLDKVPALDPRDIDDLIMGCGQPGGEAGFNIGR AVAVQLGYDFLPGTTVNRYCSSSLQTTRMAFHAIKAGEGHAFISAGVETVSRFAKGNADGWPDTKNPLYA DAMKRSDEAAAGTEEWHDPREDGALPDVYIAMGQTAENVALHTGISREDQDHWGVRSQNRAEEAINSGFF EREITPVTLPDGTVVSKDDGPRAGTTYEKISQLKPVFRPNGTVTAGNACPLNDGAAALVVMSDVRAKELG LTPLARIVSTGVSGLSPEIMGLGPIEAVKKALANANMTIGDIDLYEINEAFAVQVLGSARALGMDEDKLN VSGGAIALGHPFGMTGARITATLLNNLQTHDKTFGIETMCVGGGQGMAMVVERLS
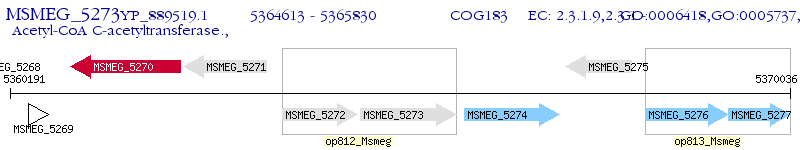
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5273 | - | - | 100% (405) | acetyl-CoA acetyltransferase |
| M. smegmatis MC2 155 | MSMEG_5199 | - | 8e-67 | 36.67% (409) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2207 | - | 1e-66 | 39.18% (416) | acetyl-CoA acetyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1103c | fadA3 | 0.0 | 84.44% (405) | acetyl-CoA acetyltransferase |
| M. gilvum PYR-GCK | Mflv_2040 | - | 0.0 | 89.88% (405) | acetyl-CoA acetyltransferase |
| M. tuberculosis H37Rv | Rv1074c | fadA3 | 0.0 | 84.44% (405) | acetyl-CoA acetyltransferase |
| M. leprae Br4923 | MLBr_02162 | fadA | 2e-62 | 33.88% (425) | acetyl-CoA acetyltransferase |
| M. abscessus ATCC 19977 | MAB_1192c | - | 0.0 | 89.63% (405) | acetyl-CoA acetyltransferase |
| M. marinum M | MMAR_4393 | fadA3 | 0.0 | 86.67% (405) | beta-ketoacyl CoA thiolase FadA3 |
| M. avium 104 | MAV_1198 | - | 0.0 | 85.68% (405) | acetyl-CoA acetyltransferase |
| M. thermoresistible (build 8) | TH_0255 | - | 1e-67 | 37.68% (406) | acetyl-CoA acetyltransferase |
| M. ulcerans Agy99 | MUL_0207 | fadA3 | 0.0 | 85.19% (405) | acetyl-CoA acetyltransferase |
| M. vanbaalenii PYR-1 | Mvan_4677 | - | 0.0 | 89.14% (405) | acetyl-CoA acetyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1103c|M.bovis_AF2122/97 -----MPEAVIVSTARSPIGRAMKGSLVGMRPDDLAVQMVRAALDKVPAL
Rv1074c|M.tuberculosis_H37Rv -----MPEAVIVSTARSPIGRAMKGSLVGMRPDDLAVQMVRAALDKVPAL
MMAR_4393|M.marinum_M MEGAVMPEAVIVSTARSPIGRAMKGSLVSMRPDDLAVQMVRAALDKVPAL
MUL_0207|M.ulcerans_Agy99 -----MPEAVIVSTARSPIGRAMKASLVSMRPDDLAVQMVRAALDKVPAL
MAV_1198|M.avium_104 -----MPEAVIVSTARSPIGRAMKGSLVSMRPDDLAVQIVRAALDKVPAL
MSMEG_5273|M.smegmatis_MC2_155 -----MPEAVIVATARSPIGRAGKGSLVEMRPDDLAAQMVKAVLDKVPAL
MAB_1192c|M.abscessus_ATCC_199 -----MPEAVIVSTARSPIGRAGKGSLVSMRPDDLAAQMVRAALDKVPAL
Mflv_2040|M.gilvum_PYR-GCK -----MPEAVIVATARSPIGRANKGSLVAMRPDDLAAQMVRAALDKVPSL
Mvan_4677|M.vanbaalenii_PYR-1 -----MPEAVIVATARSPIGRAGKGSLVSMRPDDLAAQMVRAALDKVPSL
MLBr_02162|M.leprae_Br4923 ----MSEEAFIYEAIRTPRGKQKNGSLHEVKPVSLVVGLVEELRKRNPDL
TH_0255|M.thermoresistible__bu -----MRETVIVEAVRTAVGK-RNGALADVHAADLSAVVLSELVER-AGI
*:.* : *:. *: :.:* ::. .* . :: .: . :
Mb1103c|M.bovis_AF2122/97 NPHQIDDLMMGCGLPGGESGFNIARVVAVALGY-DFLPGTTVNRYCSSSL
Rv1074c|M.tuberculosis_H37Rv NPHQIDDLMMGCGLPGGESGFNIARVVAVALGY-DFLPGTTVNRYCSSSL
MMAR_4393|M.marinum_M NPHQIDDLIMGCGLPGGESGFNIGRVVAVALGY-DFLPGTTVNRYCSSSL
MUL_0207|M.ulcerans_Agy99 NPHQIDDLIMGCGLPGGESGFNIGRVVAVALGY-DFLPGTTVNRYCSSSL
MAV_1198|M.avium_104 NPHQIDDLILGCGQPGGESGFNLARVVAVELGF-DFLPGTTVNRYCSSSL
MSMEG_5273|M.smegmatis_MC2_155 DPRDIDDLIMGCGQPGGEAGFNIGRAVAVQLGY-DFLPGTTVNRYCSSSL
MAB_1192c|M.abscessus_ATCC_199 DPKDVDDLMLGCGQPGGEAGFNIGRAVAVQLGY-DFLPGTTVNRYCSSSL
Mflv_2040|M.gilvum_PYR-GCK DPRDIDDLMMGCAQPAGEAGYNIARAVAVELGY-DFLPGTTVNRYCSSSL
Mvan_4677|M.vanbaalenii_PYR-1 DPRDIDDLMMGCAQPAGEAGYNIGRAVAIELGY-DFLPGTTVNRYCSSSL
MLBr_02162|M.leprae_Br4923 DENLISDVILGCVSPVGDQGGDIARATVLAAGLPVTAGGVQLNRFCASGL
TH_0255|M.thermoresistible__bu DPELIDDVIWGCVSQVGDQSSNIGRYAVLAAGWPETIPGTTVNRACGSSQ
: . :.*:: ** *: . ::.* ..: * *. :** *.*.
Mb1103c|M.bovis_AF2122/97 QTTRMAFHAIKAGEGDAFISAGVETVSRFAKGNSDSWPDTKNPLFDGAQE
Rv1074c|M.tuberculosis_H37Rv QTTRMAFHAIKAGEGDAFISAGVETVSRFAKGNSDSWPDTKNPLFDGAQE
MMAR_4393|M.marinum_M QTTRMAFHAIKAGEGDAFISAGVETVSRFGKGNSDSLPDTKNPLFDEAQE
MUL_0207|M.ulcerans_Agy99 QITRVAFHAIKAGEGDAFISAGVETVSRFGKGNSDSLPDTKNPLFDEAQE
MAV_1198|M.avium_104 QTSRMAFHAIKAGEGDAFISAGVETVSRFPKGSADSWPDTKNPLFSEAQE
MSMEG_5273|M.smegmatis_MC2_155 QTTRMAFHAIKAGEGHAFISAGVETVSRFAKGNADGWPDTKNPLYADAMK
MAB_1192c|M.abscessus_ATCC_199 QTTRMAFHAIKAGEGHAFISAGVETVSRFPKGTSDGWPDSHNPLYADAEA
Mflv_2040|M.gilvum_PYR-GCK QTTRMAFHAIKAGEGHAFISAGVETVSRFGKGAADGAPDSKNPIFADAQE
Mvan_4677|M.vanbaalenii_PYR-1 QTTRMAFHAIKAGEGSAFISAGVETVSRFGKGAADGAPDSKNPMFDEAQA
MLBr_02162|M.leprae_Br4923 EAVNTAAQKVRSGWDDLVLAGGVESMSRVPMGSD----------------
TH_0255|M.thermoresistible__bu QALDFAVHAVMSGQQDVVVAGGVEVMSRVPLGAAR---------------
: * : : :* .::.*** :**. *
Mb1103c|M.bovis_AF2122/97 RSAAAAAGADEWHDPRTDQKLPDIYIAMGQTAENVAIMTGISREEQDRWG
Rv1074c|M.tuberculosis_H37Rv RSAAAAAGADEWHDPRTDQKLPDIYIAMGQTAENVAIMTGISREEQDRWG
MMAR_4393|M.marinum_M RTAAAAAGAEEWHDPRADGNLPDVYIAMGQTAENVAIISGISREDQDHWG
MUL_0207|M.ulcerans_Agy99 RTAAAAAGAEEWHDPRADGNLPDVYIAMGQTAENVAIISGISREDQDHWG
MAV_1198|M.avium_104 RSAAAAEGAEEWHDPRADGKLPDIYIAMGQTAENVALLTGISREDQDHWG
MSMEG_5273|M.smegmatis_MC2_155 RSDEAAAGTEEWHDPREDGALPDVYIAMGQTAENVALHTGISREDQDHWG
MAB_1192c|M.abscessus_ATCC_199 RTATAAQGATEWHDPRNDGLTPDVYIAMGQTAENVALHTGISREDQDHWG
Mflv_2040|M.gilvum_PYR-GCK RSAKAAEGAEEWHDPREDGLLPDVYIAMGQTAENVAAFTGISREDQDHWG
Mvan_4677|M.vanbaalenii_PYR-1 RTEQGAAGATEWHDPREDGLLPDVYIAMGQTAENVANFTGISREDQDHWG
MLBr_02162|M.leprae_Br4923 -------GGAMGLDPATN--YDVMFVPQGIGADLIATIEGFSRDDVDVYA
TH_0255|M.thermoresistible__bu -----ATGMPYGPKVLER--YDNFGFNQGISAELIAERWNLSRTRLDEYS
* . . . * *: :* .:** * :.
Mb1103c|M.bovis_AF2122/97 VRSQNRAEEAIKNGFFEREITPVTLPDG-TTVSTDDGPRPGTTYEKVSEL
Rv1074c|M.tuberculosis_H37Rv VRSQNRAEEAIKNGFFEREITPVTLPDG-TTVSTDDGPRPGTTYEKVSEL
MMAR_4393|M.marinum_M VRSQNRAEEAIKSGFFEREISPVTLPDG-TTVSTDDGPRAGTTYEKVSQL
MUL_0207|M.ulcerans_Agy99 VRSQNRAEEAIKGGFFEREISPVTLPDG-TTVSTNDGPRAGTTYEKVSQL
MAV_1198|M.avium_104 VRSQNRAEEAIKSGFFEREITPVTLPDG-TTVSTDDGPRPGTTYEKIREL
MSMEG_5273|M.smegmatis_MC2_155 VRSQNRAEEAINSGFFEREITPVTLPDG-TVVSKDDGPRAGTTYEKISQL
MAB_1192c|M.abscessus_ATCC_199 VRSQNRAEKAIADGFFEREITPVTLADG-TVVAKDDGPRAGTTYEAISQL
Mflv_2040|M.gilvum_PYR-GCK VRSQNRAEEAINSGFFDREIVPVTLPDG-TVVSKDDGPRAGTSYDKISQL
Mvan_4677|M.vanbaalenii_PYR-1 VRSQNRAEEAINSGFFAREISPVTLPDG-TVVSTDDGPRAGTTYDKISQL
MLBr_02162|M.leprae_Br4923 LRSQEKAAAAWSGGYFAKSVVPVRDQNGLLILDHDEHMRPDTSKEGLAKL
TH_0255|M.thermoresistible__bu AQSHERAAAAQSSGAFKDQIVPVFTDD--AVVTEDEGIRRGTTVEKLATL
:*:::* * .* * .: ** : : :: * .*: : : *
Mb1103c|M.bovis_AF2122/97 KPAFR---------------------PNGTVTAGNACPLNDGAAAVVITS
Rv1074c|M.tuberculosis_H37Rv KPAFR---------------------PNGTVTAGNACPLNDGAAAVVITS
MMAR_4393|M.marinum_M KPVFR---------------------PNGTVTAGNACPLNDGAAAVVITS
MUL_0207|M.ulcerans_Agy99 KPVFR---------------------PNGTVTAGNACPLNDGAAAVVITS
MAV_1198|M.avium_104 KPVFR---------------------PNGTVTAGNACPLNDGAAAVVITS
MSMEG_5273|M.smegmatis_MC2_155 KPVFR---------------------PNGTVTAGNACPLNDGAAALVVMS
MAB_1192c|M.abscessus_ATCC_199 KPVFR---------------------PNGTITAGNACPLNDGAAALVVMS
Mflv_2040|M.gilvum_PYR-GCK KPVFR---------------------PNGTITAGNACPLNDGAAAVVIMS
Mvan_4677|M.vanbaalenii_PYR-1 KPVFR---------------------PNGTITAGNACPLNDGAAAVVIMS
MLBr_02162|M.leprae_Br4923 KPAFEGLAAMGGFDDVALQKYHWVEKINHVHTGGNSSGIVDGAALVMIGS
TH_0255|M.thermoresistible__bu KPAFT---------------------ENGVIHAGNSSQISDGAAALLVMT
**.* * . .**:. : **** ::: :
Mb1103c|M.bovis_AF2122/97 DTKAKELGLTPLARIVSTGVSGLSPEIMGLGPIEASKKALERAGMAITDI
Rv1074c|M.tuberculosis_H37Rv DTKAKELGLTPLARIVSTGVSGLSPEIMGLGPIEASKKALERAGMAITDI
MMAR_4393|M.marinum_M DTKAKELGLTPLARIVSTGVSGLSPEIMGLGPIEACKKALQRAGMSISDI
MUL_0207|M.ulcerans_Agy99 DTKAKELGLTPLARIVSTGVSGLSPEIMGLGPIEACKKALQRAGMSISDI
MAV_1198|M.avium_104 DTKAKELGLTPLARIVSTGVSGLSPEIMGLGPIEATKKALARAKMSVSDI
MSMEG_5273|M.smegmatis_MC2_155 DVRAKELGLTPLARIVSTGVSGLSPEIMGLGPIEAVKKALANANMTIGDI
MAB_1192c|M.abscessus_ATCC_199 DVRAKELGLTPLARIVSTGVSGLSPEIMGLGPIEAVKRALANAGKSIGDI
Mflv_2040|M.gilvum_PYR-GCK DTKAKELGLTPLARIVSTGVSGLSPEIMGLGPIEAVKKALANAKMSISDI
Mvan_4677|M.vanbaalenii_PYR-1 DTKAKELGLTPLARIVSTGVSGLSPEIMGLGPIEAVKKALANAKMSIGDI
MLBr_02162|M.leprae_Br4923 EAVGKSQGLTPRARMVATATSGADPVIMLTGPTPATRKVLDRAGLTVDDI
TH_0255|M.thermoresistible__bu AETAVTLGLTPIVRYRAGAVTGADPVLMLTGPIPATEKVLRKTGLNIDDI
. **** .* : ..:* .* :* ** * .:.* .: : **
Mb1103c|M.bovis_AF2122/97 DLVEINEAFAVQVLGSARELGIDEDKLNISGGAIALGHPFGMTGARITTT
Rv1074c|M.tuberculosis_H37Rv DLVEINEAFAVQVLGSARELGIDEDKLNISGGAIALGHPFGMTGARITTT
MMAR_4393|M.marinum_M DLVEINEAFAVQVLGSARELGIDEDKLNVSGGAIALGHPFGMTGARITTT
MUL_0207|M.ulcerans_Agy99 DLVEINEAFAVQVLGSARELGIDEDELNVSGGAIALGHPFGMTGARITTT
MAV_1198|M.avium_104 DLFEINEAFAVQVLGSARELGIDEDKLNVSGGAIALGHPFGMTGARITTT
MSMEG_5273|M.smegmatis_MC2_155 DLYEINEAFAVQVLGSARALGMDEDKLNVSGGAIALGHPFGMTGARITAT
MAB_1192c|M.abscessus_ATCC_199 DLFEINEAFAVQVLGSARALGIDEDKLNVSGGAIALGHPFGMTGARITAT
Mflv_2040|M.gilvum_PYR-GCK DLYEINEAFAVQVLGSARELGMDEDKLNVSGGAIALGHPFGMTGARITAT
Mvan_4677|M.vanbaalenii_PYR-1 DLYEINEAFAVQVLGSARELGMDEDKLNVSGGAIALGHPFGMTGARITAT
MLBr_02162|M.leprae_Br4923 DLFELNEAFASVVLKFQKDLNIPDEKLNVNGGAIAMGHPLGATGAMILGT
TH_0255|M.thermoresistible__bu GVFEVNEAFAPVPLAWQAETGADPAKLNPLGGAIALGHPLGASGAVLMTR
.: *:***** * . :** *****:***:* :** :
Mb1103c|M.bovis_AF2122/97 LLNNLQTYDKTFGLETMCVGGGQGMAMVIERLA
Rv1074c|M.tuberculosis_H37Rv LLNNLQTYDKTFGLETMCVGGGQGMAMVIERLA
MMAR_4393|M.marinum_M LLNNLQTHDKTFGLETMCVGGGQGMAMVIERLS
MUL_0207|M.ulcerans_Agy99 LLNNLQTHDKTFGLETMCVGGGQGMAMVIERLS
MAV_1198|M.avium_104 LLNNLQTHDKTFGLETMCVGGGQGMAMVIERLS
MSMEG_5273|M.smegmatis_MC2_155 LLNNLQTHDKTFGIETMCVGGGQGMAMVVERLS
MAB_1192c|M.abscessus_ATCC_199 LLNNLTTYDKTFGVETMCVGGGQGMAMVIERLA
Mflv_2040|M.gilvum_PYR-GCK LLNNLATHDKTFGIESMCVGGGQGMAMVVERLS
Mvan_4677|M.vanbaalenii_PYR-1 LLNNLTTHDKTFGIETMCVGGGQGMAMVLERLS
MLBr_02162|M.leprae_Br4923 MVDELERCNARRALVTLCIGAGMGVATIIERV-
TH_0255|M.thermoresistible__bu MVHHMRANGIRYGLQTMCEGGGTANATVVELLA
::..: . .: ::* *.* . * ::* :