For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STSAETQDNQRVDTCDLCVVGAGIAGLNALFVASRYLRRDQKIVLLDRRDRVGGMWVDTYEYVRLHQPHP MFTAGNIEWTLGREPSYLPTKTEVLDHFRHCLDVIRKQVTVDARFGWEFLSDKEFGSGPGAGVRITCRVP DGTERIIKAPRLIKAYGVRFRPNAPMKLSSTRVRSVSPDTCDVRRGDIRDSDAPVWIIGGGKTGMDTAHT LITAQTGREVNLVAGSGTYFSCRDKLFPTGIRRWGGGRSINAMAEEMCSRFDGTNEGEVADWFRREFGVW LTPQTGNFLFGVLSEAENATIAAGLREVLMDHLVDVVDRDGSTEMVLRSGARRQVEDDSWIVNCTGYLLE GDHSYEPYLSAGGSVLSIQPRSAALHLTSYSAYFLTHLFLMDKLGDLPLYEADLVELWQKHRAVAPYALF TLVQYNLGLIADTVPGKVFRECGLDFARWYPWYRRTAAGLHFMRTHRRQSEHLRRSLDTVRERFGVRCGP LIEGEARASAVAAG*
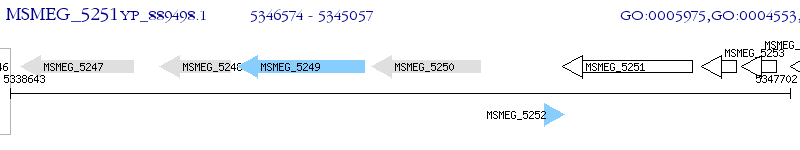
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5251 | - | - | 100% (505) | hypothetical protein MSMEG_5251 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2023 | - | 4e-07 | 26.70% (221) | hypothetical protein Mb2023 |
| M. gilvum PYR-GCK | Mflv_2222 | - | 3e-06 | 28.23% (209) | hypothetical protein Mflv_2222 |
| M. tuberculosis H37Rv | Rv2000 | - | 5e-07 | 26.70% (221) | hypothetical protein Rv2000 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0985 | - | 5e-05 | 38.33% (60) | putative monooxygenase EthA |
| M. marinum M | MMAR_2976 | - | 2e-06 | 27.01% (211) | hypothetical protein MMAR_2976 |
| M. avium 104 | MAV_4416 | - | 6e-08 | 26.64% (214) | hypothetical protein MAV_4416 |
| M. thermoresistible (build 8) | TH_2696 | - | 3e-05 | 30.07% (153) | PUTATIVE putative FAD dependent oxidoreductase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2023|M.bovis_AF2122/97 --------------------------------------------------
Rv2000|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2976|M.marinum_M --------------------------------------------------
MAV_4416|M.avium_104 --------------------------------------------------
Mflv_2222|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5251|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0985|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2696|M.thermoresistible__bu VQEIRRAVPAGGAAELSADIEHANLPSLVPVLYQLTGDRKWLGARYRPTR
Mb2023|M.bovis_AF2122/97 -----------MRPGFVGLGFGQWPVYVVRWPKLHLTPRQRKRVLHRRRL
Rv2000|M.tuberculosis_H37Rv -----------MRPGFVGLGFGQWPVYVVRWPKLHLTPRQRKRVLHRRRL
MMAR_2976|M.marinum_M --------------------------------------------------
MAV_4416|M.avium_104 --------------------------------------------------
Mflv_2222|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5251|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0985|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2696|M.thermoresistible__bu SRGMDDNDSGGFPPEVAAEIKQAAVAAVLDWEAGAKPAVPAPTGDVLLEL
Mb2023|M.bovis_AF2122/97 LTDRPISLSQIPIRTGGPMNDPWPRPTQGPAKTIETDYLVIGAGAMGMAF
Rv2000|M.tuberculosis_H37Rv LTDRPISLSQIPIRTGGPMNDPWPRPTQGPAKTIETDYLVIGAGAMGMAF
MMAR_2976|M.marinum_M ------------MREG--------EHVQAPAKAIEADYLVVGAGAMGMAF
MAV_4416|M.avium_104 ------------------------------MHTIEADYLVVGAGAMGMAF
Mflv_2222|M.gilvum_PYR-GCK ------------------------------MTGIEADYLVVGAGAMGMAF
MSMEG_5251|M.smegmatis_MC2_155 --------------------MSTSAETQDNQRVDTCDLCVVGAGIAGLNA
MAB_0985|M.abscessus_ATCC_1997 -------------------------------MTEHVDIVIVGAGISGIGM
TH_2696|M.thermoresistible__bu LELANGEHVPPEYVEFAAEDLGFIERPAPRAASPDIDVIVIGAGISGMLA
* ::*** *:
Mb2023|M.bovis_AF2122/97 TDTLITESG---ARVVMIDRACQPGGHWTT-AYPFVRLHQPSAYYGVN--
Rv2000|M.tuberculosis_H37Rv TDTLITESG---ARVVMIDRACQPGGHWTT-AYPFVRLHQPSAYYGVN--
MMAR_2976|M.marinum_M TDTLITESD---ARVVIVDRAHAPGGHWTT-AYPFVRLHQPSAYYGVN--
MAV_4416|M.avium_104 IDTLVTETAASQARVVVVDRHHAPGGHWTI-AYPFVRLHQPSAFYGVN--
Mflv_2222|M.gilvum_PYR-GCK VDTLLAETRDAGTTVVLVDENHQPGGHWNT-VYPFVRLHQPSAYYGVN--
MSMEG_5251|M.smegmatis_MC2_155 LFVASRYLR-RDQKIVLLDRRDRVGGMWVD-TYEYVRLHQPHPMFTAGNI
MAB_0985|M.abscessus_ATCC_1997 ACRVTRELP--GKKFVVLEARERIGGTWD--LFRYPGVRSDSDMFTLG--
TH_2696|M.thermoresistible__bu VIQLRQAGF---ERIVVLEKNDDVGGCWLENRYPGAGVDTPSYLYSYS--
.*::: ** * : : : .
Mb2023|M.bovis_AF2122/97 SRALGNN---TIDLVGWNQGLNELAPVGEICAYFDAVLQQQLLPTGRVDY
Rv2000|M.tuberculosis_H37Rv SRALGNN---TIDLVGWNQGLNELAPVGEICAYFDAVLQQQLLPTGRVDY
MMAR_2976|M.marinum_M SRPLGNN---TIDAVGWNQGLNELAPVGEICAYFDAVMQQQFLPTGRVEY
MAV_4416|M.avium_104 SLRLGGD---SIDQIGWNRGLYELATADEICAYYDRVMRTRLLPTGRVRY
Mflv_2222|M.gilvum_PYR-GCK SEPLGDEE--AIDRSGFNEGFFELASGHEVCAYYDHVMRNHLLPTGRLTY
MSMEG_5251|M.smegmatis_MC2_155 EWTLGREPSYLPTKTEVLDHFRHCLDVIRKQVTVDARFGWEFLSDKEFGS
MAB_0985|M.abscessus_ATCC_1997 -----YNFRPWTSTKGIADGTSIREYVTETAREFGVTEKVRFG--HKVVG
TH_2696|M.thermoresistible__bu -------FTQRDWATHFAKRDEVENYLRRVADEYRVRDAIRFN--TEVVS
. .: ..
Mb2023|M.bovis_AF2122/97 FPMSEYLGDGRFRTLAGTEYVVTVNRRIVDATYLRAVVPSMRPAPYSVAP
Rv2000|M.tuberculosis_H37Rv FPMSEYLGDGRFRTLAGTEYVVTVNRRIVDATYLRAVVPSMRPAPYSVAP
MMAR_2976|M.marinum_M FPMSEYLGDGRFRTLGGAEHQVTVSRRIVDATYLRAVVPSMRPTPYPVAP
MAV_4416|M.avium_104 FPDSEYRGDGRFRTSAGADHTVEVTRRIVDATYMHVSVPAMRPPPYTVGD
Mflv_2222|M.gilvum_PYR-GCK LPMTRYLGDNRIRTLDGAEQTVTVRRRTVDATYLLTVVQSMRSAPFSVAD
MSMEG_5251|M.smegmatis_MC2_155 GPGAGVR--ITCRVPDGTERIIKAPRLIKAYGVRFRPNAPMKLSSTRVRS
MAB_0985|M.abscessus_ATCC_1997 AEWSSERGLWTLQVERAGETVEFTTQFLLGCTGYYRYDEGFTPKFEGIED
TH_2696|M.thermoresistible__bu AEFDPRSARWTVRTDRGDE---LSARILISATGVLNRPK--IPSIPGRES
:. . : :
Mb2023|M.bovis_AF2122/97 GVDCVAPNELP--KLG-TRDRYVVVGAGKTGMDVCLWLLRNDVC------
Rv2000|M.tuberculosis_H37Rv GVDCVAPNELP--KLG-TRDRYVVVGAGKTGMDVCLWLLRNDVC------
MMAR_2976|M.marinum_M GIDCIAPNDLP--KFT-GKDRYVIVGAGKTAMDVCLWLLRNSVD------
MAV_4416|M.avium_104 GVDCVPPNELP--RRP-GYQRYVVVGAGKTGIDTCLWLLGQGVE------
Mflv_2222|M.gilvum_PYR-GCK GVEVVAPNDLP--RRAPHHDHYTVVGGGKTGMDCCLWLLRNGVP------
MSMEG_5251|M.smegmatis_MC2_155 VSPDTCDVRRG--DIRDSDAPVWIIGGGKTGMDTAHTLITAQTGREVNL-
MAB_0985|M.abscessus_ATCC_1997 FAGQVVHPQHWPEDLDYSGKRVVVIGSGATAMTLVPAMAGTAGHVTMLQR
TH_2696|M.thermoresistible__bu FEGPAFHTAEWPDGLDLSGKRVAVIGTGASAMQVVPAIAGEVESLWVFQR
::* * :.: :
Mb2023|M.bovis_AF2122/97 -PDKLT------------------WIMPRDSWLIDRATLQPGPTFVRQFR
Rv2000|M.tuberculosis_H37Rv -PDKLT------------------WIMPRDSWLIDRATLQPGPTFVRQFR
MMAR_2976|M.marinum_M -PGQLS------------------WIKPRDAWLIDRATLQPGPTFIKQFR
MAV_4416|M.avium_104 -PDRLT------------------WIMPRDSWLLDRATMQPGALFAEQIK
Mflv_2222|M.gilvum_PYR-GCK -ADAVR------------------WVRPRDSWLLNRAHIQPGEQFAPQLR
MSMEG_5251|M.smegmatis_MC2_155 VAGSGT------------------YFSCRDKLFPTGIRRWGGGRSINAMA
MAB_0985|M.abscessus_ATCC_1997 SPTYVV------------------SLPSTDALAVALQGKLPASLAYPIVK
TH_2696|M.thermoresistible__bu SPQWIAPNNVYFSAIDPAVHRLMARVPFYARWYRARLAWNFNDRVHPSLQ
. . .
Mb2023|M.bovis_AF2122/97 ES-----YGATLEAIGAATSTDDLFDRL-----ETAGTLLRIDPSVRPSM
Rv2000|M.tuberculosis_H37Rv ES-----YGATLEAIGAATSTDDLFDRL-----ETAGTLLRIDPSVRPSM
MMAR_2976|M.marinum_M DS-----YGITLEAIGRATSIRDLFDRL-----EKAGTLLRLDPAVRPTM
MAV_4416|M.avium_104 HS-----FTAQLRAINEAGSVAELFSRL-----EAAGLLLRIDPAVRPTM
Mflv_2222|M.gilvum_PYR-GCK DA-----MAARMQAISEAASVEHLFTLL-----EDAEVLLRLDPSVWPTM
MSMEG_5251|M.smegmatis_MC2_155 EE-----MCSRFDGTNEGEVADWFRREFGVWLTPQTGNFLFGVLSEAENA
MAB_0985|M.abscessus_ATCC_1997 WKNQMVSFISYQTSRRDPERMKGILRALLKRQLPNFDLDKHFTPKYNPWD
TH_2696|M.thermoresistible__bu IDPDWPHFDRSINAANDG-HRRVFTRYIEEQLSDRPDLIEKVLPDYPPFG
. : :
Mb2023|M.bovis_AF2122/97 YRCATVSHLELEQLRRIRD-------IVRMGHVQRIEPTTIVLDGGSVPA
Rv2000|M.tuberculosis_H37Rv YRCATVSHLELEQLRRIRD-------IVRMGHVQRIEPTTIVLDGGSVPA
MMAR_2976|M.marinum_M YRCATVSQPEFDQLRRIED-------VVRMGHVQRIDSTEIVLDGGSVPC
MAV_4416|M.avium_104 YRCATVTRLELEQLRRITD-------VVRLGHVRSIEPGAMVLDGGAVAV
Mflv_2222|M.gilvum_PYR-GCK YHCAIVSQRELEYLRRIED-------VVRLGHIEHAEPDRVRLTGATIPA
MSMEG_5251|M.smegmatis_MC2_155 TIAAGLREVLMDHLVDVVDRDGSTEMVLRSGARRQVEDDSWIVNCTGYLL
MAB_0985|M.abscessus_ATCC_1997 QRLCVVPDSDLFRALRKGT------ASIVTDRITRFTPKGILLESG-EEL
TH_2696|M.thermoresistible__bu KRMLLDNG--WYAALRRPN------VHLVTEAVTRIGPSSVYGAGGTEAE
:
Mb2023|M.bovis_AF2122/97 TPTALYIDCTADGAPQRPAKPV-----FDADHLTLQAVRGCQQVFSAAFI
Rv2000|M.tuberculosis_H37Rv TPTALYIDCTADGAPQRPAKPV-----FDADHLTLQAVRGCQQVFSAAFI
MMAR_2976|M.marinum_M SASALYVDCTADGAPQRPAKPV-----FDGDHLTLQAVRGCQQVFSAAFT
MAV_4416|M.avium_104 DGRALYLDCTADGAEKRPATPI-----FTPGRITLQSVRGCQQIFSAALI
Mflv_2222|M.gilvum_PYR-GCK TP-SLYVDCTTQGLPRPPSVPV-----FDGDRITLQSVRSCQQVFSSAFI
MSMEG_5251|M.smegmatis_MC2_155 EGDHSYEPYLSAGGSVLSIQPR-----SAALHLTSYSAYFLTHLFLMDKL
MAB_0985|M.abscessus_ATCC_1997 EADIVVTATGLNVQLAGGLRPI-----VDGVQLNPADSVSYKGLMLTGLP
TH_2696|M.thermoresistible__bu VDVVVFATGFHTHRYLYPMRIAGRSGRTLADTWGPENAHAYLGITVPDFP
:
Mb2023|M.bovis_AF2122/97 AHV-----EFAYEDDAVKNELCTPIPHPDCDLDWMRLMHSDLGNFQRWLN
Rv2000|M.tuberculosis_H37Rv AHV-----EFAYEDDAVKNELCTPIPHPDCDLDWMRLMHSDLGNFQRWLN
MMAR_2976|M.marinum_M AHV-----ELAYDGDEVKNELCTPIPHPDSDLDWMRLTHSDLRNFQRWLD
MAV_4416|M.avium_104 AHV-----EAGYGDDARRNELCVPLPHPDTDVDWLRLALADYTNQLRWFD
Mflv_2222|M.gilvum_PYR-GCK AHA-----ETTFSDDDTRNALCVPVVHPDAPIDWLRHMSEDNAASMRWIR
MSMEG_5251|M.smegmatis_MC2_155 GDLPLY--EADLVELWQKHRAVAPYALFTLVQYNLGLIADTVPGKVFREC
MAB_0985|M.abscessus_ATCC_1997 NFIFTF--GYTNASWTLRADLVSQYTCRLLKYMDKEGQTVVWPVEPDTAE
TH_2696|M.thermoresistible__bu NLFILCGPGTALGHGGSFITVAECQVRYVVDLLVTMAERGLRTVEVRPEV
Mb2023|M.bovis_AF2122/97 DPDLTDWLSSARLN----LLADLLPPLS-HKPRVRERVVSMFQK-RLGTA
Rv2000|M.tuberculosis_H37Rv DPDLTDWLSSARLN----LLADLLPPLS-HKPRVRERVVSMFQK-RLGTA
MMAR_2976|M.marinum_M DVELTDWLSSARLN----LLADLLPPLS-HKPRVRDRVVSMFRS-KLNIA
MAV_4416|M.avium_104 DPELMTWLSGARLDLFGHLIGHLLPSAS-AKPRVRARILGIAKS-VFAAT
Mflv_2222|M.gilvum_PYR-GCK DPELMTWLRTARLN----AARDLFPVFPPGKERVRDKAMGALAV-ALRKT
MSMEG_5251|M.smegmatis_MC2_155 GLDFARWYPWYRRT----AAGLHFMRTHRRQSEHLRRSLDTVRE-RFGVR
MAB_0985|M.abscessus_ATCC_1997 RLPFLEDLVSGYVTRAENAMPHQVPLAPWRSYQNYFRELPVLKYGKVADK
TH_2696|M.thermoresistible__bu EADYTRRHDDAHAK-----MLWSHPGMTNWYRNEQGRVVATMPWRIVDYW
. : : .
Mb2023|M.bovis_AF2122/97 GDQLAKLLDAA--TATTEQR---------
Rv2000|M.tuberculosis_H37Rv GDQLAKLLDAA--TATTEQR---------
MMAR_2976|M.marinum_M SEQLEKLLSDHGHDGASQQQ---------
MAV_4416|M.avium_104 AARLGELLAAEAALAAS------------
Mflv_2222|M.gilvum_PYR-GCK NEELAALMAGAS-----------------
MSMEG_5251|M.smegmatis_MC2_155 CGPLIEGEARASAVAAG------------
MAB_0985|M.abscessus_ATCC_1997 GVRFSSGAAGSATSGRGNASHEAPVAIGR
TH_2696|M.thermoresistible__bu RMTRCAALEDYRCERDDAAVAVDA-----