For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PTRQQPSFPRGTALLVAGALFMEILDATILAPAIPLIAESFGVAAVDVNVAISAYLVTVAVLIPATGWMA DRFGVRRVFISAIAVFTLASVGCALSVSLPMLVAMRVAQGVGGAMMVPVGRLAVLRSSDKADLVRAIALL TWPALAAPVVAPVLGGAIATLGDWRWIFAVNVPIGLIGILFALRLIHGDAPARPRSLDWFGLLTLGSGIA ATLLALENIRVSGTHWSLVGSFGVAAVLLLAVALRRLLTVPNPLVQLRVLRVQTLRITVSAGSLYRMVIT AVPFLLPLQFQLQFGWTPLAAGAMVAALFAGNLTIKPITTPLMRRFGIRTVLLVNGAASVACFGLLAVLR PGLPVVLMAVILYISGALRSIGFTAYNSLAFSDVEGDDLTHGNTLNASVQELASGLGIAFAALLLSQLTS YSLTFLVLGAVLALTLVETVRLPKSAGSHVSGHR*
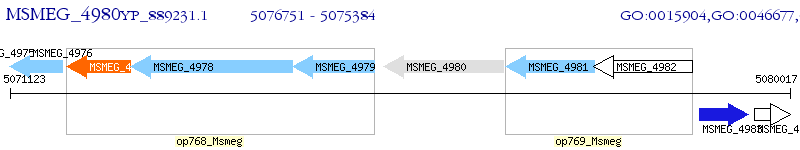
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4980 | - | - | 100% (455) | major facilitator superfamily protein MFS_1 |
| M. smegmatis MC2 155 | MSMEG_5670 | - | 7e-32 | 26.99% (415) | efflux membrane protein |
| M. smegmatis MC2 155 | MSMEG_0827 | - | 2e-30 | 30.07% (459) | major facilitator superfamily protein MFS_1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0805c | emrB | 3e-43 | 27.82% (417) | multidrug resistance integral membrane efflux protein EmrB |
| M. gilvum PYR-GCK | Mflv_2018 | - | 0.0 | 73.41% (455) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv0783c | emrB | 3e-43 | 27.82% (417) | multidrug resistance integral membrane efflux protein EmrB |
| M. leprae Br4923 | MLBr_01562 | - | 3e-25 | 26.82% (425) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_1396 | - | 2e-32 | 31.30% (345) | EmrB/QacA family drug resistance transporter |
| M. marinum M | MMAR_2966 | - | 3e-48 | 30.32% (432) | integral membrane drug efflux protein |
| M. avium 104 | MAV_0730 | - | 5e-49 | 29.73% (444) | EmrB efflux protein |
| M. thermoresistible (build 8) | TH_2729 | - | 9e-31 | 29.83% (362) | PUTATIVE putative drug resistance transporter, EmrB/QacA |
| M. ulcerans Agy99 | MUL_0542 | - | 3e-41 | 27.93% (401) | multidrug resistance integral membrane efflux protein |
| M. vanbaalenii PYR-1 | Mvan_1284 | - | 7e-42 | 33.56% (432) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4980|M.smegmatis_MC2_155 ----------------------------------MPTRQQPSFPRG----
Mflv_2018|M.gilvum_PYR-GCK ----------------------------------MPAASEPTFPRG----
Mb0805c|M.bovis_AF2122/97 ----------------------MLGNAMVEACPAEGDAPVPITPAGRPRS
Rv0783c|M.tuberculosis_H37Rv ----------------------MLGNAMVEACPAEGDAPVPITPAGRPRS
MAV_0730|M.avium_104 MRSPSAITELAAAGAGESGLFDMLSNAMDEAPYAAAKTP-PHAPTGQPGA
MMAR_2966|M.marinum_M -------------------------------MRRAGTPRRPRSPDTHPDN
MUL_0542|M.ulcerans_Agy99 --------------------------------------------------
Mvan_1284|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_1396|M.abscessus_ATCC_1997 --------------------------------------MRPEYPRP----
TH_2729|M.thermoresistible__bu --------------------------------------------------
MLBr_01562|M.leprae_Br4923 -----------MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTS
MSMEG_4980|M.smegmatis_MC2_155 ----------------TALLVAGA--LFMEILDATILAPAIPLIAESFGV
Mflv_2018|M.gilvum_PYR-GCK ----------------MALLVAGA--MFMEILDATILAPAIPAIAATFGV
Mb0805c|M.bovis_AF2122/97 -GQRSYPDRLDVGLLRTAGVCVLA--SVMAHVDVTVVSVAQRTFVADFGS
Rv0783c|M.tuberculosis_H37Rv -GQRSYPDRLDVGLLRTAGVCVLA--SVMAHVDVTVVSVAQRTFVADFGS
MAV_0730|M.avium_104 -TEREYPDKLDAALLRISGVCILA--TVMAILDVTVVSVAQRTFIDQFSS
MMAR_2966|M.marinum_M RHQHSYPDRLDAGLWRIAGVCVLG--SIMTMVDTSVVTVAQRTFVDTFGS
MUL_0542|M.ulcerans_Agy99 ---------------MTTATCLML--PVMVTLDTTVVNVAQRTFIEEFSS
Mvan_1284|M.vanbaalenii_PYR-1 -------------MKWILAAVATA--LFCVQIDYFAMNLALPRIAQEFGE
MAB_1396|M.abscessus_ATCC_1997 --------------WITLWVVLFG--LFMILLDSTIVSVANPAIKAGFGA
TH_2729|M.thermoresistible__bu ----------------VLVALILT--MALVAMDTTILATAVPQVVGDLG-
MLBr_01562|M.leprae_Br4923 KYPTLLPSGRFPSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSL
:* * . :.
MSMEG_4980|M.smegmatis_MC2_155 AAVDVNVAISAYLVTVAVLIPATGWMADRFGVRRVFISAIAVFTLASVGC
Mflv_2018|M.gilvum_PYR-GCK AAVDVNIAISSYLLTAAVLIPASGWIADRFGIRPVFLTAIAVFTIASIGC
Mb0805c|M.bovis_AF2122/97 TQAVVAWTMTGYMLALATVIPTAGWAADRFGTRRLFMGSVLAFTLGSLLC
Rv0783c|M.tuberculosis_H37Rv TQAVVAWTMTGYMLALATVIPTAGWAADRFGTRRLFMGSVLAFTLGSLLC
MAV_0730|M.avium_104 SQAVVAWTMTGYTLALATVIPITGWAADRFGTKRLFIGSVVAFMLGSLLC
MMAR_2966|M.marinum_M TQAVVAWTITGYTLALAAVVPLAGWAADRFGTKRMFMGSILVFTLSSLLC
MUL_0542|M.ulcerans_Agy99 TQAVVAWTSTGYTLSLASVIPLTGWTANRLGTKRLVMGSVLLFTLGSLLC
Mvan_1284|M.vanbaalenii_PYR-1 SPSDLQWVISVYMLALGAFMVPAGRMGDIFGRRRALLTGVALFGGASVAC
MAB_1396|M.abscessus_ATCC_1997 DYSSTVWVTSAYLLGYAVPLLITGRLGDQFGPRSMYLSGLAVFTAASLWC
TH_2729|M.thermoresistible__bu GFDQVGWVFSGYLLAQTVTIPIYGKLADLYGRRPILIFGVVAFLIGSALS
MLBr_01562|M.leprae_Br4923 SDAGRSWGITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLC
: * : : * .: * : : .: * .* .
MSMEG_4980|M.smegmatis_MC2_155 ALSVSLPMLVAMRVAQGVGGAMMVPVGRLAVLRSSDK-ADLVRAIALLTW
Mflv_2018|M.gilvum_PYR-GCK ALSATLPMLVAMRVLQGVGGAMMVPVGRLAVLRFSAK-ADLVRAIALLTW
Mb0805c|M.bovis_AF2122/97 AVAPNILLLIIFRVVQGFGGGMLTPVSFAILAREAGP-KRLGRVMAVVGI
Rv0783c|M.tuberculosis_H37Rv AVAPNILLLIIFRVVQGFGGGMLTPVSFAILAREAGP-KRLGRVMAVVGI
MAV_0730|M.avium_104 ALAANILQLIVFRVVQGIGGGMLLPLGFMILTREAGP-RRLGRLMSILSI
MMAR_2966|M.marinum_M AIAPNIALLIASRVVQGLGGGMLAPLALTIVNREAGP-KRVGRVMAVLGI
MUL_0542|M.ulcerans_Agy99 AMASSITLLVAFRAAQGLGGGILIPLQLIILARAAGP-ARLGRVLTISMI
Mvan_1284|M.vanbaalenii_PYR-1 ALAPSVSLLVAFRAVQGLGAALIFPVSVSVLTNAYDA-AKAGRAIGLAYG
MAB_1396|M.abscessus_ATCC_1997 GLAGSIGWLIAARVVQGIGAAMLTPQTLTVVQRVFPP-ERRGTAMGVWGA
TH_2729|M.thermoresistible__bu AASWNMATLAIFRAIQGLGAGAISATVQTVAGDLYTV-AERGRIQGYLAS
MLBr_01562|M.leprae_Br4923 AVAWDEVTLVIARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAA
. : * * **.*.. .
MSMEG_4980|M.smegmatis_MC2_155 PALAAPVVAPVLGGAIATLGDWRWIFAVNVPIGLIGILFALRLIHGDAPA
Mflv_2018|M.gilvum_PYR-GCK PALTAPVLAPVLGGALATLGSWRWIFIVNIPIGIVGFVLALKLIRGGPAP
Mb0805c|M.bovis_AF2122/97 PMLLGPVGGPILGGWLIGAYGWRWIFLVNLPVGLSALVLAAIVFPRDRPA
Rv0783c|M.tuberculosis_H37Rv PMLLGPVGGPILGGWLIGAYGWRWIFLVNLPVGLSALVLAAIVFPRDRPA
MAV_0730|M.avium_104 PMLLAPIAGPILGGWLIDTSSWRWIFLINVPIGLLTVALAAVVFPRDHPA
MMAR_2966|M.marinum_M PGVLAPAFGPALGGWLIDSYSWQWIFWVNLPVGVVAVGLAAVVFPRDTPA
MUL_0542|M.ulcerans_Agy99 SVLMAPIAGPILGGWLIDAFGWQWIFLINLPIGALTLILAGLVLPGDDSL
Mvan_1284|M.vanbaalenii_PYR-1 IAGLGNAAGPLVGGLLTGTVGWRWIFWLNVPLALASFVLGARHITESFDE
MAB_1396|M.abscessus_ATCC_1997 VAGIATLVGPVAGGLLVDGWGWQWIFYVNVPVGVLGIILGAIYIPRMPTH
TH_2729|M.thermoresistible__bu VWGISAVIAPALGGFFAQYLTWRWIFLVNIPIGVFALYLIARDLHEDVVR
MLBr_01562|M.leprae_Br4923 MTAIGSVMGLVVGGALTEVS-WRLAFLVNVPIGLVMIYLARTALHETHKE
. . ** : *: * :*:*:. . : :
MSMEG_4980|M.smegmatis_MC2_155 RP-RSLDWFGLLTLGSGIAATLLALENIRVSG--THWSLVGSFGVAAVLL
Mflv_2018|M.gilvum_PYR-GCK TP-RPFDWRGLLLLGAGIGAALVALEHIRVTG--TAWVLVGAGLVGAVAL
Mb0805c|M.bovis_AF2122/97 AS-ENFDYMGLLLLSPGLATFLFGVSSSPARGTMADRHVLIPAITGLALI
Rv0783c|M.tuberculosis_H37Rv AS-ENFDYMGLLLLSPGLATFLFGVSSSPARGTMADRHVLIPAITGLALI
MAV_0730|M.avium_104 RS-ETFDAVGVLLLSPGLATFLFAVSSIPGRGTVADRHVLIPAAMGLTLI
MMAR_2966|M.marinum_M PS-ETFDVVGMLLLSPGLPAFLYGMSEIPIYGTVADRHVWVPAGIGIALI
MUL_0542|M.ulcerans_Agy99 PA-ESLDLVGMALLSPALVLLLYGLSALPARGTIGDPQVWLPTTVGLILI
Mvan_1284|M.vanbaalenii_PYR-1 TAPRRIDITGLALVTAGIALFTLAFDRAPDWGWLSVPTLG--VFTGAVLA
MAB_1396|M.abscessus_ATCC_1997 RH--RLDLLGMALSAVGMFAFVFALQE--GESFQWAPGVWAMMAAGLVVL
TH_2729|M.thermoresistible__bu RP-HRIDYLGAGLVLVSAGLFILGLLSG-GVHWSWTSPTSVVVFTGAALA
MLBr_01562|M.leprae_Br4923 RM--KLDAAGAILATLACTAAVFAFSMGPEKGWISLTTIG--SGLVALVA
:* * . ..
MSMEG_4980|M.smegmatis_MC2_155 LAVALRRLLTVPNPLVQLRVLRVQTLRITVSAGSLYRMVITAVPFLLPLQ
Mflv_2018|M.gilvum_PYR-GCK LAAAVWHLRRTPTPLVRLAVLRVRTLRITVVGGSVYRLVITAVPFLLPLL
Mb0805c|M.bovis_AF2122/97 AAFVAHSWYRTEHPLIDMRLFQNRAVAQANMTMTVLSLGLFGSFLLLPSY
Rv0783c|M.tuberculosis_H37Rv AAFVAHSWYRTEHPLIDMRLFQNRAVAQANMTMTVLSLGLFGSFLLLPSY
MAV_0730|M.avium_104 AGFVGHAWHRADHPLIDLRLFRNPVLTHANVTMLVFATAFFGAGLLLPSY
MMAR_2966|M.marinum_M VGFMFHALYRADKPLIDLRLLTNRALTLANVAMFLYIVSTFGAGVLFPSY
MUL_0542|M.ulcerans_Agy99 GGFALHALHRGDHALIDLRLLKNRAVAAANATRFMFAVTFFGSCLLFPAY
Mvan_1284|M.vanbaalenii_PYR-1 LAVFVAVENRVRRPLIDLSLFRAGRFTILVVAGTVANVAYAVVIFLSTLE
MAB_1396|M.abscessus_ATCC_1997 GVFVWWQSANRGEPLIPLRLFADRNFSLAGVGIASMSFCVISTGLPMMFY
TH_2729|M.thermoresistible__bu GVALLVVESRAAEPVLPLSMLTRRLTAPSFAATATAGMIVIGLSVYLPNW
MLBr_01562|M.leprae_Br4923 AIAFAVVERTAENPVVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLY
.:: : :: .
MSMEG_4980|M.smegmatis_MC2_155 FQLQFGWTPLAAGAMVAALFAGNLTIKPITTPLMRRFGIRTVLLVNGAAS
Mflv_2018|M.gilvum_PYR-GCK FQLEFGWSAFTAGLMVAALFIGNLTIKPLTTPLMRRFGIKRVLLVNGVVS
Mb0805c|M.bovis_AF2122/97 LQQVLHQSPMQSGVHIIPQGLGAMLAMPIAGAMMDRRGPAKIVLVGIMLI
Rv0783c|M.tuberculosis_H37Rv LQQVLHQSPMQSGVHIIPQGLGAMLAMPIAGAMMDRRGPAKIVLVGIMLI
MAV_0730|M.avium_104 FQQVLHQTPMQAGVHMIPQGLGAMLTMRLTGPLVDRQGPGKVVLVGIALI
MMAR_2966|M.marinum_M FQQLLDHTPLQAGMSLLPRGIGAALAVPLAGALVDRRGARGVLVIGVTLI
MUL_0542|M.ulcerans_Agy99 FQQVLGKTPLQSGLLLIPQTLAAAAVMPIVGRLMEKRGPRDVVLMGTALT
Mvan_1284|M.vanbaalenii_PYR-1 LQQVRGLDPLAAGLVFLGPSTGAAIGGVMAGRLATGRSP-----VAVMGN
MAB_1396|M.abscessus_ATCC_1997 TQLALGFSPTKAALTQAPTAIVSGILAPLAGWLVDRVRPSILIGGGIALM
TH_2729|M.thermoresistible__bu GQTVLGLSPVAAGFVLAIMSITWPISSGFSARLYLRIGFRDTALVGASCA
MLBr_01562|M.leprae_Br4923 IQDILGYSALRAGIGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLL
* . :. . :
MSMEG_4980|M.smegmatis_MC2_155 VACFG--LLAVLRPGLPVVLMAVILYISGALRSIGFTAYNSLAFSDVEGD
Mflv_2018|M.gilvum_PYR-GCK VAWFG--VLALLEPGTPVALIAAALYVSGALRSIGFTAYNSLAFADVDGD
Mb0805c|M.bovis_AF2122/97 AAGLGTFAFGVARQADYLPILPTGLAIMGMGMGCSMMPLSGAAVQTLAPH
Rv0783c|M.tuberculosis_H37Rv AAGLGTFAFGVARQADYLPILPTGLAIMGMGMGCSMMPLSGAAVQTLAPH
MAV_0730|M.avium_104 TAGLGAFAFGVARQAPYLPTLLAGLAITGLGMGCTMMPLSVASVQALAPH
MMAR_2966|M.marinum_M ATGMGVFAFGVATQRDYLPMLLIGLTILGMGMGCTRMPLVAVAMQSLAPN
MUL_0542|M.ulcerans_Agy99 VVGMGIFIYGMSREHVHLPALLIGLGTFGVGTAAMMVPVSWSAVHMLNSS
Mvan_1284|M.vanbaalenii_PYR-1 TSVVAALCLAALAAGGPWWFHVAALSACGFSLGLVYAFTTVATQAVVRPE
MAB_1396|M.abscessus_ATCC_1997 IASTLWWTLLMRPETQMWQLMLPAAGIG-AAMACIWGPLATTATRNLPPS
TH_2729|M.thermoresistible__bu VAAGTG--LVLLGPAPPVWQPVVYTALMGAGMGLLFSPLIVGLQNTVGWD
MLBr_01562|M.leprae_Br4923 MVAMLYGWAFMNRGVPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFD
. :
MSMEG_4980|M.smegmatis_MC2_155 DLTHGNTLNASVQELASGLGIAFAALLLS---------------------
Mflv_2018|M.gilvum_PYR-GCK ELTHANTLNATVQEMGAGLGIAVAALLLT---------------------
Mb0805c|M.bovis_AF2122/97 QIARGSTLISVNQQVGGSIGTALMSVLLTYQFNHSEIIATAKKVALTPES
Rv0783c|M.tuberculosis_H37Rv QIARGSTLISVNQQVGGSIGTALMSVLLTYQFNHSEIIATAKKVALTPES
MAV_0730|M.avium_104 QIARGTTLMSVSHQVGGSMGTALMSMILTNQFNRSPNIVAANKLAALHQK
MMAR_2966|M.marinum_M QIARGSTLIKVNQQMAAAVGTALMSVILTSQLNNGENASGASKAEILREH
MUL_0542|M.ulcerans_Agy99 EVAHGSTLFNVNHNVAASVGAALD--------------VGRTHQPIQRQR
Mvan_1284|M.vanbaalenii_PYR-1 RAGEAAGVTLTALVTAGGVGIAMAATIFEVLQ------------------
MAB_1396|M.abscessus_ATCC_1997 VAGAGSGVYNTLRQVGGVVGSSAMGALMTARLTATMPAMPPGSGSTPEAA
TH_2729|M.thermoresistible__bu QRGTVTGGLMFSRFLGQSIGAAGFGAVANAVLRRYEPDTSAAAMG-----
MLBr_01562|M.leprae_Br4923 RIGPVSAIALMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQL
. : :
MSMEG_4980|M.smegmatis_MC2_155 -----------------------------QLTSYSLTFLVLGAVLALTLV
Mflv_2018|M.gilvum_PYR-GCK -----------------------------VLGSYPWTFLALGAILALTLI
Mb0805c|M.bovis_AF2122/97 GAGRGAAVDPSSLPRQ---TNFAAQLLHDLSHAYAVVFVIATALVVSTLI
Rv0783c|M.tuberculosis_H37Rv GAGRGAAVDPSSLPRQ---TNFAAQLLHDLSHAYAVVFVIATALVVSTLI
MAV_0730|M.avium_104 AAAGGTPIDQSAIPRQSLAPGFGGNVLHDLSHAYTAVFVIAVALVVCTII
MMAR_2966|M.marinum_M RP--GVPLDQP--------------VLHELSRAYSVVFVVAVVLAVLAFI
MUL_0542|M.ulcerans_Agy99 --------------------------------------------------
Mvan_1284|M.vanbaalenii_PYR-1 ---------------------------------REGMSPAAAIAVILGAV
MAB_1396|M.abscessus_ATCC_1997 LPPG---------------------LRAPFSAAMAQSMLLGIAALALGLV
TH_2729|M.thermoresistible__bu -------------------------------AASHAVFVGLLVAAVATVV
MLBr_01562|M.leprae_Br4923 R---------------------------ALDHGYTYGLLWVAGAVVIVGV
MSMEG_4980|M.smegmatis_MC2_155 ETVRLPKSAGSHVSGHR---------------------------------
Mflv_2018|M.gilvum_PYR-GCK ETLRLPGSAASHVTGHR---------------------------------
Mb0805c|M.bovis_AF2122/97 PAAFLPKQQASHRRAPLLSA------------------------------
Rv0783c|M.tuberculosis_H37Rv PAAFLPKQQASHRRAPLLSA------------------------------
MAV_0730|M.avium_104 PASFLPKKPAAETAGK----------------------------------
MMAR_2966|M.marinum_M PLAFLPKWSALTALSQTSSLERTPEPDAAPHGRQSPPEDTRPGAGRNSSS
MUL_0542|M.ulcerans_Agy99 --------------------------------------------------
Mvan_1284|M.vanbaalenii_PYR-1 SALLLPAGAAVLLIKAPA--------------------------------
MAB_1396|M.abscessus_ATCC_1997 AALFMRPGGSQVGAVPLDDVVSVED-------------------------
TH_2729|M.thermoresistible__bu LLLTVPRRFPTHHVAVVD--------------------------------
MLBr_01562|M.leprae_Br4923 AALFIGYTPEQVAYAQEVKEAVDAGEL-----------------------
MSMEG_4980|M.smegmatis_MC2_155 ----------
Mflv_2018|M.gilvum_PYR-GCK ----------
Mb0805c|M.bovis_AF2122/97 ----------
Rv0783c|M.tuberculosis_H37Rv ----------
MAV_0730|M.avium_104 ----------
MMAR_2966|M.marinum_M NDLTPSGPDG
MUL_0542|M.ulcerans_Agy99 ----------
Mvan_1284|M.vanbaalenii_PYR-1 ----------
MAB_1396|M.abscessus_ATCC_1997 ----------
TH_2729|M.thermoresistible__bu ----------
MLBr_01562|M.leprae_Br4923 ----------