For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTSLALRSPVALDPPTSARRGLRSFLADPGLVCGSVLVGLVALAAFSAPLVAALVGHGPNDQFPDDTLDD SGLPITGGKGFLFGADGSGRDVFVRTLYGARISLAIGVPATTIAMVVGVVVGLAGGYFGGRIDAALSELT NVALAFPFLVTALSVVTLNRGTAGTTLVDPIFVVIGIIALFSWTYFARLVRNSVIELKARPFVLAAVGSG SSHTRVLVREILPNIAPTVIIYWAVQLPTNIVAEASLSYLGVGIKPPTPSWGTMIANAQDSALYQVQPLM LIAPAGALFVTVLGFNILSTRLRSRFDPLSAR
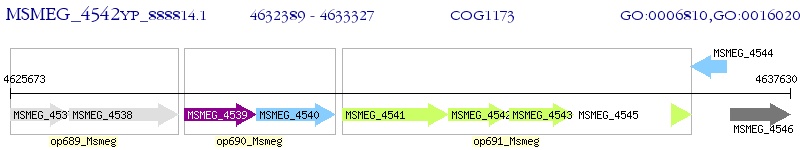
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4542 | - | - | 100% (312) | oligopeptide transport integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_0642 | - | 1e-42 | 35.49% (293) | ABC transporter permease |
| M. smegmatis MC2 155 | MSMEG_6867 | - | 3e-39 | 35.61% (278) | oligopeptide ABC transporter integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3688c | dppC | 3e-39 | 35.59% (281) | peptide ABC transporter transmembrane protein |
| M. gilvum PYR-GCK | Mflv_2759 | - | 7e-36 | 33.33% (297) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3664c | dppC | 3e-39 | 35.59% (281) | peptide ABC transporter transmembrane protein |
| M. leprae Br4923 | MLBr_01123 | - | 1e-29 | 33.06% (245) | putative binding-protein dependent transport protein |
| M. abscessus ATCC 19977 | MAB_0428 | - | 4e-36 | 36.00% (225) | peptide ABC transporter DppC |
| M. marinum M | MMAR_5152 | dppC | 6e-42 | 36.07% (280) | dipeptide-transport integral membrane protein ABC |
| M. avium 104 | MAV_0466 | dppC | 5e-40 | 37.41% (278) | ABC transporter, permease protein DppC |
| M. thermoresistible (build 8) | TH_2705 | - | 7e-36 | 34.27% (286) | PUTATIVE binding-protein-dependent transport systems inner |
| M. ulcerans Agy99 | MUL_4238 | dppC | 2e-42 | 36.90% (271) | dipeptide-transport integral membrane protein ABC |
| M. vanbaalenii PYR-1 | Mvan_0437 | - | 4e-47 | 39.39% (297) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2759|M.gilvum_PYR-GCK ------------MTTTDPQS------TRLSSWRLLALNPVTLISALVLIA
TH_2705|M.thermoresistible__bu ------------MTADDLTGGEPDAVSRVAAWRLLAANPVTLLSAALLAL
Mvan_0437|M.vanbaalenii_PYR-1 ------MVAVPEAPPGVPESQVQGRNPWYLAWLRLRRNKVALGCGLLFVL
MLBr_01123|M.leprae_Br4923 ---------MTLTEGLSISEKTNFASRKTLVLRRFAHNRTASVSLMLLVL
Mb3688c|M.bovis_AF2122/97 -----------------------------------------MIAAALILL
Rv3664c|M.tuberculosis_H37Rv -----------------------------------------MIAAALILL
MAV_0466|M.avium_104 ----------------MAERIRARGGFWRETWRRLRRRPKFIGAGLLILV
MMAR_5152|M.marinum_M --------------------MAELGGFWFDILGGLRRRPKFVIAAVLIVF
MUL_4238|M.ulcerans_Agy99 --------------------MAELRGFWFDILGGLRRRPKFVIAAVLIVF
MAB_0428|M.abscessus_ATCC_1997 MSDPVNTPGPVSGPDATAESLAEQADLWGNAWKYLRRSGFFITGSILLTI
MSMEG_4542|M.smegmatis_MC2_155 ----------MTSLALRSPVALDPPTSARRGLRSFLADPGLVCGSVLVGL
:.
Mflv_2759|M.gilvum_PYR-GCK VAVVAVTANWIIPYGVN-------------------DVDVPN--ALAP-P
TH_2705|M.thermoresistible__bu LVAVAVTAPWLAPYGVN-------------------DINVPE--ALQP-P
Mvan_0437|M.vanbaalenii_PYR-1 IVAVCLAAPLWADYVAHTGPNDNHITDTVLIDGVETDIVAPDGTPLGPGL
MLBr_01123|M.leprae_Br4923 LFVACYLLPSLLPYSYD----------------------DLDFNALLQPP
Mb3688c|M.bovis_AF2122/97 ILVVAAFPSLFT---AADPT-------------------YADPSQSMLAP
Rv3664c|M.tuberculosis_H37Rv ILVVAAFPSLFT---AADPT-------------------YADPSQSMLAP
MAV_0466|M.avium_104 ILAVALFPALFT---AADPT-------------------YADPAQSLLPP
MMAR_5152|M.marinum_M ILVVAAFPSLFT---GIDPS-------------------YADPSQSMLSP
MUL_4238|M.ulcerans_Agy99 ILFVAAFPSLFT---GIDPS-------------------YADPSQSMLSP
MAB_0428|M.abscessus_ATCC_1997 LVLMALAPQLFTF--GKDPR-------------------ACDLYQARKGI
MSMEG_4542|M.smegmatis_MC2_155 VALAAFSAPLVAALVGHGPND-------------QFPDDTLDDSGLPITG
: . :
Mflv_2759|M.gilvum_PYR-GCK SGSHWFGTDELGRDVLSRVLVATQASMRIAVVSVAFAVIVGVAVGVIAGY
TH_2705|M.thermoresistible__bu SAAHWFGTDELGRDVLSRVLVAIQASLRVAAVSVAFAAAVGILVGLLAGY
Mvan_0437|M.vanbaalenii_PYR-1 RGRYLLGADQNGRDVMVRLLYGGRTSIYIGLAAAAVTTVLAVVVALLAGY
MLBr_01123|M.leprae_Br4923 NTRHWLGTNALGQDLLAQTLRGMQKSMLIGVCVALISTSIAATVGSIAGY
Mb3688c|M.bovis_AF2122/97 SAAHWFGTDLQGHDIYSRTVYGARASVTVGLGATLAVFVVGGALGALAGF
Rv3664c|M.tuberculosis_H37Rv SAAHWFGTDLQGHDIYSRTVYGARASVTVGLGATLAVFVVGGALGALAGF
MAV_0466|M.avium_104 SRTHWFGTDLQGHDVYARTVYGARASVTVGLGAAAIVFVVGGALGALAGF
MMAR_5152|M.marinum_M STAHWFGTDLQGHDIYARTVYGARASVVVGLGATAAAFIAGVTLGALAGY
MUL_4238|M.ulcerans_Agy99 STAHWFGTDLQGHDIYARTVYGARASVVVGLGATAAAFIAGVTLGALAGY
MAB_0428|M.abscessus_ATCC_1997 SAAHWLGTDLQGCDYYANVVYGARVSLTIGLIAMVGVLIIGVIVGALAGY
MSMEG_4542|M.smegmatis_MC2_155 GKGFLFGADGSGRDVFVRTLYGARISLAIGVPATTIAMVVGVVVGLAGGY
. :*:: * * . : . : *: :. . :. .*:
Mflv_2759|M.gilvum_PYR-GCK RGGWLDTVLMRVVDVMFAFPVLLL-------------ALAVVAILG----
TH_2705|M.thermoresistible__bu RGGWLDTVLMRLVDVLFAFPVLLL-------------ALAIVAILG----
Mvan_0437|M.vanbaalenii_PYR-1 YRGWIDAVLSRVLDVIWAFPVLLLGIALGTALAIGGLKLGVIAIAG----
MLBr_01123|M.leprae_Br4923 FGRWRDRVLMWVVDLLLVVPSFVLIA-----------IMSPQTKSS----
Mb3688c|M.bovis_AF2122/97 YGSWIDAVVSRVTDVFLGLPLLLAAIV----------LMQVMHHRT----
Rv3664c|M.tuberculosis_H37Rv YGSWIDAVVSRVTDVFLGLPLLLAAIV----------LMQVMHHRT----
MAV_0466|M.avium_104 YGGWIDAVVSRVTDVFFGLPLLLAAIV----------LMQVLHHRT----
MMAR_5152|M.marinum_M YGGWLDAVVSRITDIFFGLPLLLAAIV----------LMQVMHHRT----
MUL_4238|M.ulcerans_Agy99 YGGWLDAVVSRITDIFFGLPLLLAAIV----------LMQVMHHRT----
MAB_0428|M.abscessus_ATCC_1997 FGGIADSALSRLTDVFYGIPTILGGMI----------LLSVVGHRT----
MSMEG_4542|M.smegmatis_MC2_155 FGGRIDAALSELTNVALAFPFLVTALS----------VVTLNRGTAGTTL
* .: : :: .* :: :
Mflv_2759|M.gilvum_PYR-GCK -PGVTTTILAIGIVYTPIFARVARASTLGVRTEPYVSVSRAMGAGDLYIL
TH_2705|M.thermoresistible__bu -PGMTTTMLAIGIVFTPIFARVARAGTLGVRVEPFVQASHTMGTGHAYIL
Mvan_0437|M.vanbaalenii_PYR-1 -DSIWIPILIIGLVYVPYMARPLRGEILALREKEFVEAAVAQGMGPLRIM
MLBr_01123|M.leprae_Br4923 -DNIMLLVLLLAGFGWMVSSRMVRGMTMSLRKSEFIRAGKYMGVSSRRMI
Mb3688c|M.bovis_AF2122/97 ---VWTVIAILALFGWPQVARIARGAVLEVRASDYVLAAKALGLNRFQIL
Rv3664c|M.tuberculosis_H37Rv ---VWTVIAILALFGWPQVARIARGAVLEVRASDYVLAAKALGLNRFQIL
MAV_0466|M.avium_104 ---VWTVIAILALFGWPQVARIARGAVLAVRASDYVLAAKALGMSRFQIL
MMAR_5152|M.marinum_M ---VWTVIAILALFGWPQIARIARSSVLEVRGSDFVLAAKALGMSRFQIL
MUL_4238|M.ulcerans_Agy99 ---VWTVIAILALFGWPQIARIARSSVLEVRGSDFVLAAKALGMSRFQIL
MAB_0428|M.abscessus_ATCC_1997 ---VWSVAGALIAFGWMTSMRLVRSSVMSIKQSDYVQAAIALGAPTWRIL
MSMEG_4542|M.smegmatis_MC2_155 VDPIFVVIGIIALFSWTYFARLVRNSVIELKARPFVLAAVGSGSSHTRVL
: : . * * : :: :: .. * ::
Mflv_2759|M.gilvum_PYR-GCK RRHILPNIAGPLIVQTSLSLAFAILSEAALSFLGLGIQPPQPSLGRMIFD
TH_2705|M.thermoresistible__bu RRHILPNIAAPLIVQTSLSLAFAILAEAALSFLGLGIQPPEPSLGRMIFD
Mvan_0437|M.vanbaalenii_PYR-1 VGELLPNIVSTIIVFFTLNIANNMLLESALSFLGAGVRPPNASWGTMIAD
MLBr_01123|M.leprae_Br4923 VGHVVPNVASILIIDATLNVGSAILAETGLSFLGFGIKPPDVSLGTLIAN
Mb3688c|M.bovis_AF2122/97 LRHALPNAVGPVIAVATVALGIFIVTEATLSYLGVGLPTSVVSWGGDINV
Rv3664c|M.tuberculosis_H37Rv LRHALPNAVGPVIAVATVALGIFIVTEATLSYLGVGLPTSVVSWGGDINV
MAV_0466|M.avium_104 IRHALPNALGPVIAVATIALGLFIVTEATLSYLGVGLPPSVVSWGGDINL
MMAR_5152|M.marinum_M LRHALPNAIGPVIAVATVALGIFIVTEATLSYLGVGLPPTVVSWGGDINV
MUL_4238|M.ulcerans_Agy99 LRHALPNAIGPVIAVATVALGIFIVTEATLSYLGVGLPPTVVSWGDDINV
MAB_0428|M.abscessus_ATCC_1997 LRHILPNALAPVLVYATIAVGSFIAAEATLTYMGIGLELPEISWGLQIND
MSMEG_4542|M.smegmatis_MC2_155 VREILPNIAPTVIIYWAVQLPTNIVAEASLSYLGVGIKPPTPSWGTMIAN
. :** :: :: : : *: *:::* *: . * * *
Mflv_2759|M.gilvum_PYR-GCK SQG--FVTMAWWMAVFPGAAIFLIVLAFNLFGDGLRDVLDPKQRTTIEAR
TH_2705|M.thermoresistible__bu SQG--FVTLAWWMAVFPGAAIVAAVLACNLFGDGLRDVLDPKQRTVIETQ
Mvan_0437|M.vanbaalenii_PYR-1 GYQ--MIYTAPHLTIVPGLMIVVTVLSLNVFGDGLRDALDPKARIRIEAN
MLBr_01123|M.leprae_Br4923 GTQ--SATTFPWVFLFPAGVLVVILVCANLTGDGLRDAFDPVSKHLRR--
Mb3688c|M.bovis_AF2122/97 AQT--RLRSGSPILFYPAGALAITVLAFMMMGDALRDALDPASRAWRA--
Rv3664c|M.tuberculosis_H37Rv AQT--RLRSGSPILFYPAGALAITVLAFMMMGDALRDALDPASRAWRA--
MAV_0466|M.avium_104 AQT--RLRAGSPILFYPAGALAATVLAFMMMGDALRDALDPASRAWRA--
MMAR_5152|M.marinum_M AQM--RLRAGSPILFYPAGALAVTVLAFMMMGDALRDALDPASRAWRA--
MUL_4238|M.ulcerans_Agy99 AQM--RLRAGSPILFYPAGALAVTVLAFMMMGDALRDALDPASRAWRA--
MAB_0428|M.abscessus_ATCC_1997 GQS--RFASTPRLVIVPALFLSAAVLGFIMVGDALRDALDPRRR------
MSMEG_4542|M.smegmatis_MC2_155 AQDSALYQVQPLMLIAPAGALFVTVLGFNILSTRLRSRFDPLSAR-----
. : . *. : :: : . **. :**
Mflv_2759|M.gilvum_PYR-GCK RLQR-------------
TH_2705|M.thermoresistible__bu RRARQRPGRGRRAGSAW
Mvan_0437|M.vanbaalenii_PYR-1 DAGRH------------
MLBr_01123|M.leprae_Br4923 -----------------
Mb3688c|M.bovis_AF2122/97 -----------------
Rv3664c|M.tuberculosis_H37Rv -----------------
MAV_0466|M.avium_104 -----------------
MMAR_5152|M.marinum_M -----------------
MUL_4238|M.ulcerans_Agy99 -----------------
MAB_0428|M.abscessus_ATCC_1997 -----------------
MSMEG_4542|M.smegmatis_MC2_155 -----------------