For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDVAEPVTVAGPVPATGHSPWYLAWTRLRRNKVALFNGGVFILIVLFCLAAPLWSEHVAHTLPNENHITD QVLIDGVETYVVSPDGTPIGPGLHGRYLLGADQNGRDVMVRLMYGGRTSIYIGLTAALVTTVLAVVIGLL AGFYRGWIDTVLSRIMDVMWAFPVLLLGIALGTALALGGFKLGVIHVAGDSLWIPILIIGCVYVPYMARP IRGEVLALRQKEFIEAAVAMGKGPVRIMVTELLPNIISTIIVFFTLNIANNMLLESALSFLGAGVRPPNA SWGTMIADGYQTIYTAPHLTIVPGLMIVITVLSLNVFGDGLREALDPKAKIRLEH*
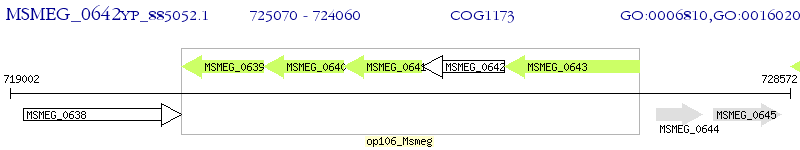
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0642 | - | - | 100% (336) | ABC transporter permease |
| M. smegmatis MC2 155 | MSMEG_6867 | - | 4e-50 | 39.92% (238) | oligopeptide ABC transporter integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_1086 | - | 1e-46 | 35.08% (325) | ABC transporter permease protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1313c | oppC | 2e-35 | 32.77% (235) | oligopeptide-transport integral membrane protein ABC |
| M. gilvum PYR-GCK | Mflv_2759 | - | 3e-45 | 38.75% (240) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv1282c | oppC | 2e-35 | 32.77% (235) | oligopeptide-transport integral membrane protein ABC |
| M. leprae Br4923 | MLBr_01123 | - | 5e-36 | 32.34% (235) | putative binding-protein dependent transport protein |
| M. abscessus ATCC 19977 | MAB_0428 | - | 1e-37 | 29.71% (340) | peptide ABC transporter DppC |
| M. marinum M | MMAR_4137 | oppC | 1e-36 | 30.84% (308) | oligopeptide-transport integral membrane protein ABC |
| M. avium 104 | MAV_1432 | oppC | 2e-37 | 35.47% (234) | ABC transporter, permease protein OppC |
| M. thermoresistible (build 8) | TH_2705 | - | 4e-44 | 32.80% (311) | PUTATIVE binding-protein-dependent transport systems inner |
| M. ulcerans Agy99 | MUL_4000 | oppC | 1e-36 | 30.84% (308) | oligopeptide-transport integral membrane protein ABC |
| M. vanbaalenii PYR-1 | Mvan_0437 | - | 1e-157 | 83.96% (318) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0642|M.smegmatis_MC2_155 ---MTDVAEPVT---VAGPVPATGHSPWYLAWTRLRRNKVALFNGGVFIL
Mvan_0437|M.vanbaalenii_PYR-1 ---MVAVPEAPP---GVPESQVQGRNPWYLAWLRLRRNKVALGCGLLFVL
Mb1313c|M.bovis_AF2122/97 --------------------MTEFASRRTLVVRRFLRNRAAVASLAALLL
Rv1282c|M.tuberculosis_H37Rv --------------------MTEFASRRTLVVRRFLRNRAAVASLAALLL
MMAR_4137|M.marinum_M ---MTDQA-----------TQVEFTSRRTLVLRRFLRNRAAVASLAVLVV
MUL_4000|M.ulcerans_Agy99 ---MTDQA-----------TQVEFTSRRTLVLRRFLRNRAAVASLAVLVV
MAV_1432|M.avium_104 ---MTAQANVDGAHGFSGHEVAGFASRRTLVLRRFGRNRLAVTSLTLLVL
MLBr_01123|M.leprae_Br4923 ---MTLTEG------LSISEKTNFASRKTLVLRRFAHNRTASVSLMLLVL
Mflv_2759|M.gilvum_PYR-GCK ---MTTTD---------PQS------TRLSSWRLLALNPVTLISALVLIA
TH_2705|M.thermoresistible__bu ---MTADD---------LTGGEPDAVSRVAAWRLLAANPVTLLSAALLAL
MAB_0428|M.abscessus_ATCC_1997 MSDPVNTPGPVSGPDATAESLAEQADLWGNAWKYLRRS-GFFITGSILLT
: . :
MSMEG_0642|M.smegmatis_MC2_155 IVLFCLAAPLWSEHVAHTLPNENHITDQVLIDGVETYVVSPDGTPIGPGL
Mvan_0437|M.vanbaalenii_PYR-1 IVAVCLAAPLWADYVAHTGPNDNHITDTVLIDGVETDIVAPDGTPLGPGL
Mb1313c|M.bovis_AF2122/97 LFVSAYALPPLLPYSYDDLDFNALLQP----------------------P
Rv1282c|M.tuberculosis_H37Rv LFVSAYALPPLLPYSYDDLDFNALLQP----------------------P
MMAR_4137|M.marinum_M LFVGCYALPSVLPYSYEDLDFDALLQP----------------------P
MUL_4000|M.ulcerans_Agy99 LFVGCYALPSVLPYSYEDLDFDALLQP----------------------P
MAV_1432|M.avium_104 LFVGCYTLPAVLPYSYQDLDFDALLQP----------------------P
MLBr_01123|M.leprae_Br4923 LFVACYLLPSLLPYSYDDLDFNALLQP----------------------P
Mflv_2759|M.gilvum_PYR-GCK VAVVAVTANWIIPYGVNDVDVPNALAP----------------------P
TH_2705|M.thermoresistible__bu LVAVAVTAPWLAPYGVNDINVPEALQP----------------------P
MAB_0428|M.abscessus_ATCC_1997 ILVLMALAPQLFTFGKDPRACDLYQARKG--------------------I
: . .
MSMEG_0642|M.smegmatis_MC2_155 HGRYLLGADQNGRDVMVRLMYGGRTSIYIGLTAALVTTVLAVVIGLLAGF
Mvan_0437|M.vanbaalenii_PYR-1 RGRYLLGADQNGRDVMVRLLYGGRTSIYIGLAAAAVTTVLAVVVALLAGY
Mb1313c|M.bovis_AF2122/97 GTKHWLGTNALGQDLLAQTLRGMQKSMLIGVCVAVISTGIAATVGAISGY
Rv1282c|M.tuberculosis_H37Rv GTKHWLGTNALGQDLLAQTLRGMQKSMLIGVCVAVISTGIAATVGAISGY
MMAR_4137|M.marinum_M STRHWLGTNALGQDLLAQTLRGMQKSMLIGVFVAVISTGIAATVGAVSGY
MUL_4000|M.ulcerans_Agy99 STRHWLGTNALGQDLLAQTLRGMQKSMLIGVFVAVISTGIAATVGAVSGY
MAV_1432|M.avium_104 NARHWLGTNALGQDLLAQILRGMQKSMLIGVCVAVISTGIAATVGSIAGY
MLBr_01123|M.leprae_Br4923 NTRHWLGTNALGQDLLAQTLRGMQKSMLIGVCVALISTSIAATVGSIAGY
Mflv_2759|M.gilvum_PYR-GCK SGSHWFGTDELGRDVLSRVLVATQASMRIAVVSVAFAVIVGVAVGVIAGY
TH_2705|M.thermoresistible__bu SAAHWFGTDELGRDVLSRVLVAIQASLRVAAVSVAFAAAVGILVGLLAGY
MAB_0428|M.abscessus_ATCC_1997 SAAHWLGTDLQGCDYYANVVYGARVSLTIGLIAMVGVLIIGVIVGALAGY
: :*:: * * . : . : *: :. :. :. ::*:
MSMEG_0642|M.smegmatis_MC2_155 YRGWIDTVLSRIMDVMWAFPVLLLGIALGTALALGGFKLGVIHVAGDSLW
Mvan_0437|M.vanbaalenii_PYR-1 YRGWIDAVLSRVLDVIWAFPVLLLGIALGTALAIGGLKLGVIAIAGDSIW
Mb1313c|M.bovis_AF2122/97 FGGWRDRTLMWVVDLLLVVPSFILIAIVTPRTKNS-----------ANIM
Rv1282c|M.tuberculosis_H37Rv FGGWRDRTLMWVVDLLLVVPSFILIAIVTPRTKNS-----------ANIM
MMAR_4137|M.marinum_M FGGWRDRALMWLVDLLLVVPSFILIAIVTPRTKNS-----------TNMM
MUL_4000|M.ulcerans_Agy99 FGGWRDRALMWLVDLLLVVPSFILIAIVTPRTKNS-----------TNMM
MAV_1432|M.avium_104 FGGWRDRVLMWLVDLLLVVPSFILIAIVTPRTKNS-----------ANIL
MLBr_01123|M.leprae_Br4923 FGRWRDRVLMWVVDLLLVVPSFVLIAIMSPQTKSS-----------DNIM
Mflv_2759|M.gilvum_PYR-GCK RGGWLDTVLMRVVDVMFAFPVLLLALAVVAILGPG-----------VTTT
TH_2705|M.thermoresistible__bu RGGWLDTVLMRLVDVLFAFPVLLLALAIVAILGPG-----------MTTT
MAB_0428|M.abscessus_ATCC_1997 FGGIADSALSRLTDVFYGIPTILGGMILLSVVGHR------------TVW
* .* : *:: .* :: : . .
MSMEG_0642|M.smegmatis_MC2_155 IPILIIGCVYVPYMARPIRGEVLALRQKEFIEAAVAMGKGPVRIMVTELL
Mvan_0437|M.vanbaalenii_PYR-1 IPILIIGLVYVPYMARPLRGEILALREKEFVEAAVAQGMGPLRIMVGELL
Mb1313c|M.bovis_AF2122/97 FLVLLLAGFGWMISSRMVRGMTMSLREREFIRAARYMGVSSRRIIVGHVV
Rv1282c|M.tuberculosis_H37Rv FLVLLLAGFGWMISSRMVRGMTMSLREREFIRAARYMGVSSRRIIVGHVV
MMAR_4137|M.marinum_M FLVLLLAGFGWMISSRMVRGMTMSLREREFIRAARYMGVSSRRIITGHVI
MUL_4000|M.ulcerans_Agy99 FLVLLLAGFGWMISSRMVRGMTMSLREREFIRAARYMGVSSRRIITGHVI
MAV_1432|M.avium_104 MLVLLLAGFGWMVSSRMVRGMTMSLREREFIRAARYMGVSSRRIIVGHVV
MLBr_01123|M.leprae_Br4923 LLVLLLAGFGWMVSSRMVRGMTMSLRKSEFIRAGKYMGVSSRRMIVGHVV
Mflv_2759|M.gilvum_PYR-GCK ILAIGIVYT--PIFARVARASTLGVRTEPYVSVSRAMGAGDLYILRRHIL
TH_2705|M.thermoresistible__bu MLAIGIVFT--PIFARVARAGTLGVRVEPFVQASHTMGTGHAYILRRHIL
MAB_0428|M.abscessus_ATCC_1997 SVAGALIAFGWMTSMRLVRSSVMSIKQSDYVQAAIALGAPTWRILLRHIL
: * *. :.:: :: .. * :: .::
MSMEG_0642|M.smegmatis_MC2_155 PNIISTIIVFFTLNIANNMLLESALSFLGAGVRPPNASWGTMIADGYQTI
Mvan_0437|M.vanbaalenii_PYR-1 PNIVSTIIVFFTLNIANNMLLESALSFLGAGVRPPNASWGTMIADGYQMI
Mb1313c|M.bovis_AF2122/97 PNVASILIIDAALNVAAAILAETGLSFLGFGIQPPDVSLGTLIADGTASA
Rv1282c|M.tuberculosis_H37Rv PNVASILIIDAALNVAAAILAETGLSFLGFGIQPPDVSLGTLIADGTASA
MMAR_4137|M.marinum_M PNVASILIIDAALNVAAAILAETGLSFLGFGIQPPDVSLGTLIADGTQSA
MUL_4000|M.ulcerans_Agy99 PNVASILIIDAALNVAAAILAETGLSFLGFGIQPPDVSLGTLIADGTQSA
MAV_1432|M.avium_104 PNVASILIIDAALNVASAILAETGLSFLGFGVQPPDVSLGTLIANGTQSA
MLBr_01123|M.leprae_Br4923 PNVASILIIDATLNVGSAILAETGLSFLGFGIKPPDVSLGTLIANGTQSA
Mflv_2759|M.gilvum_PYR-GCK PNIAGPLIVQTSLSLAFAILSEAALSFLGLGIQPPQPSLGRMIFDSQGFV
TH_2705|M.thermoresistible__bu PNIAAPLIVQTSLSLAFAILAEAALSFLGLGIQPPEPSLGRMIFDSQGFV
MAB_0428|M.abscessus_ATCC_1997 PNALAPVLVYATIAVGSFIAAEATLTYMGIGLELPEISWGLQINDGQSRF
** . ::: :: :. : *: *:::* *:. *: * * * :.
MSMEG_0642|M.smegmatis_MC2_155 YTAPHLTIVPGLMIVITVLSLNVFGDGLREALDPKAKIRLEH--------
Mvan_0437|M.vanbaalenii_PYR-1 YTAPHLTIVPGLMIVVTVLSLNVFGDGLRDALDPKARIRIEANDAGRH--
Mb1313c|M.bovis_AF2122/97 TAFPWVFLFPASILVLILVCANLTGDGLRDALDPASRSLRRGVR------
Rv1282c|M.tuberculosis_H37Rv TAFPWVFLFPASILVLILVCANLTGDGLRDALDPASRSLRRGVR------
MMAR_4137|M.marinum_M TTFPWVFLFPAGVLVLILVCANLTGDGLRDALDPASKTLRSGDR------
MUL_4000|M.ulcerans_Agy99 TTFPWVFLFPAGVLVLILVCANLTGDGLRDALDPASKTLRSGDR------
MAV_1432|M.avium_104 TTFPWVFLFPAGVLVLILVCANLTGDGLRDALDPGSGPARGGRR------
MLBr_01123|M.leprae_Br4923 TTFPWVFLFPAGVLVVILVCANLTGDGLRDAFDPVSKHLRR---------
Mflv_2759|M.gilvum_PYR-GCK TMAWWMAVFPGAAIFLIVLAFNLFGDGLRDVLDPKQRTTIEARRLQR---
TH_2705|M.thermoresistible__bu TLAWWMAVFPGAAIVAAVLACNLFGDGLRDVLDPKQRTVIETQRRARQRP
MAB_0428|M.abscessus_ATCC_1997 ASTPRLVIVPALFLSAAVLGFIMVGDALRDALDPRRR-------------
: :.*. : :: : **.**:.:**
MSMEG_0642|M.smegmatis_MC2_155 ----------
Mvan_0437|M.vanbaalenii_PYR-1 ----------
Mb1313c|M.bovis_AF2122/97 ----------
Rv1282c|M.tuberculosis_H37Rv ----------
MMAR_4137|M.marinum_M ----------
MUL_4000|M.ulcerans_Agy99 ----------
MAV_1432|M.avium_104 ----------
MLBr_01123|M.leprae_Br4923 ----------
Mflv_2759|M.gilvum_PYR-GCK ----------
TH_2705|M.thermoresistible__bu GRGRRAGSAW
MAB_0428|M.abscessus_ATCC_1997 ----------