For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAERIRARGGFWRETWRRLRRRPKFIGAGLLILVILAVALFPALFTAADPTYADPAQSLLPPSRTHWFGT DLQGHDVYARTVYGARASVTVGLGAAAIVFVVGGALGALAGFYGGWIDAVVSRVTDVFFGLPLLLAAIVL MQVLHHRTVWTVIAILALFGWPQVARIARGAVLAVRASDYVLAAKALGMSRFQILIRHALPNALGPVIAV ATIALGLFIVTEATLSYLGVGLPPSVVSWGGDINLAQTRLRAGSPILFYPAGALAATVLAFMMMGDALRD ALDPASRAWRA
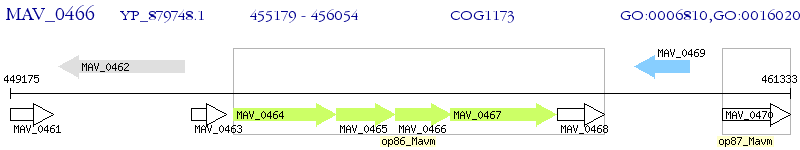
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0466 | dppC | - | 100% (291) | ABC transporter, permease protein DppC |
| M. avium 104 | MAV_1432 | oppC | 2e-34 | 30.60% (268) | ABC transporter, permease protein OppC |
| M. avium 104 | MAV_3419 | - | 1e-32 | 38.29% (222) | ABC transporter permease protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3688c | dppC | 1e-133 | 88.64% (264) | peptide ABC transporter transmembrane protein |
| M. gilvum PYR-GCK | Mflv_2759 | - | 3e-47 | 36.63% (273) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3664c | dppC | 1e-133 | 88.64% (264) | peptide ABC transporter transmembrane protein |
| M. leprae Br4923 | MLBr_01123 | - | 3e-37 | 31.64% (275) | putative binding-protein dependent transport protein |
| M. abscessus ATCC 19977 | MAB_0428 | - | 2e-73 | 47.74% (287) | peptide ABC transporter DppC |
| M. marinum M | MMAR_5152 | dppC | 1e-135 | 81.98% (283) | dipeptide-transport integral membrane protein ABC |
| M. smegmatis MC2 155 | MSMEG_6867 | - | 1e-48 | 39.63% (270) | oligopeptide ABC transporter integral membrane protein |
| M. thermoresistible (build 8) | TH_2705 | - | 7e-44 | 36.40% (272) | PUTATIVE binding-protein-dependent transport systems inner |
| M. ulcerans Agy99 | MUL_4238 | dppC | 1e-134 | 81.56% (282) | dipeptide-transport integral membrane protein ABC |
| M. vanbaalenii PYR-1 | Mvan_3775 | - | 1e-48 | 37.73% (273) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2759|M.gilvum_PYR-GCK ------------MTTTDPQS------TRLSSWRLLALNPVTLISALVLIA
Mvan_3775|M.vanbaalenii_PYR-1 ------------MTTTDPND------TRLSSWRLLAGNPVTLVSALVLVA
TH_2705|M.thermoresistible__bu ------------MTADDLTGGEPDAVSRVAAWRLLAANPVTLLSAALLAL
MSMEG_6867|M.smegmatis_MC2_155 ------------------------------MHRVLKAR-MAPLALVVLAL
Mb3688c|M.bovis_AF2122/97 -----------------------------------------MIAAALILL
Rv3664c|M.tuberculosis_H37Rv -----------------------------------------MIAAALILL
MAV_0466|M.avium_104 ----------------MAERIRARGGFWRETWRRLRRRPKFIGAGLLILV
MMAR_5152|M.marinum_M --------------------MAELGGFWFDILGGLRRRPKFVIAAVLIVF
MUL_4238|M.ulcerans_Agy99 --------------------MAELRGFWFDILGGLRRRPKFVIAAVLIVF
MAB_0428|M.abscessus_ATCC_1997 MSDPVNTPGPVSGPDATAESLAEQADLWGNAWKYLRRSGFFITGSILLTI
MLBr_01123|M.leprae_Br4923 ------------MTLTEGLSISEKTNFASRKTLVLRRFAHNRTASVSLML
. :
Mflv_2759|M.gilvum_PYR-GCK VAVVAVTANWII---PYGVNDVDVPNALAPP--SGSHWFGTDELGRDVLS
Mvan_3775|M.vanbaalenii_PYR-1 VTVVAVTANWLA---PYGVNDVDVPNALAPP--SASHWFGTDELGRDVLS
TH_2705|M.thermoresistible__bu LVAVAVTAPWLA---PYGVNDINVPEALQPP--SAAHWFGTDELGRDVLS
MSMEG_6867|M.smegmatis_MC2_155 VVLCAVLAPVLS---PFDPNAQDLLTRLSPPQRTGGHLLGTDNLGRDVLS
Mb3688c|M.bovis_AF2122/97 ILVVAAFPSLFT---AADPTYADPSQSMLAP--SAAHWFGTDLQGHDIYS
Rv3664c|M.tuberculosis_H37Rv ILVVAAFPSLFT---AADPTYADPSQSMLAP--SAAHWFGTDLQGHDIYS
MAV_0466|M.avium_104 ILAVALFPALFT---AADPTYADPAQSLLPP--SRTHWFGTDLQGHDVYA
MMAR_5152|M.marinum_M ILVVAAFPSLFT---GIDPSYADPSQSMLSP--STAHWFGTDLQGHDIYA
MUL_4238|M.ulcerans_Agy99 ILFVAAFPSLFT---GIDPSYADPSQSMLSP--STAHWFGTDLQGHDIYA
MAB_0428|M.abscessus_ATCC_1997 LVLMALAPQLFTF--GKDPRACDLYQARKGI--SAAHWLGTDLQGCDYYA
MLBr_01123|M.leprae_Br4923 LVLLFVACYLLPSLLPYSYDDLDFNALLQPP--NTRHWLGTNALGQDLLA
: : . : . * :**: * * :
Mflv_2759|M.gilvum_PYR-GCK RVLVATQASMRIAVVSVAFAVIVGVAVGVIAGYRGGWLDTVLMRVVDVMF
Mvan_3775|M.vanbaalenii_PYR-1 RVLVAVQASMRIAAVSVALAMVVGVAVGVVAGYRGGWLDTVLMRVVDVMF
TH_2705|M.thermoresistible__bu RVLVAIQASLRVAAVSVAFAAAVGILVGLLAGYRGGWLDTVLMRLVDVLF
MSMEG_6867|M.smegmatis_MC2_155 RLIYGARISLFVGLAVALISGTIGALVGIVAGYRGGWADRVLMRLADVQL
Mb3688c|M.bovis_AF2122/97 RTVYGARASVTVGLGATLAVFVVGGALGALAGFYGSWIDAVVSRVTDVFL
Rv3664c|M.tuberculosis_H37Rv RTVYGARASVTVGLGATLAVFVVGGALGALAGFYGSWIDAVVSRVTDVFL
MAV_0466|M.avium_104 RTVYGARASVTVGLGAAAIVFVVGGALGALAGFYGGWIDAVVSRVTDVFF
MMAR_5152|M.marinum_M RTVYGARASVVVGLGATAAAFIAGVTLGALAGYYGGWLDAVVSRITDIFF
MUL_4238|M.ulcerans_Agy99 RTVYGARASVVVGLGATAAAFIAGVTLGALAGYYGGWLDAVVSRITDIFF
MAB_0428|M.abscessus_ATCC_1997 NVVYGARVSLTIGLIAMVGVLIIGVIVGALAGYFGGIADSALSRLTDVFY
MLBr_01123|M.leprae_Br4923 QTLRGMQKSMLIGVCVALISTSIAATVGSIAGYFGRWRDRVLMWVVDLLL
. : . : *: :. . :* :**: * * .: :.*:
Mflv_2759|M.gilvum_PYR-GCK AFP-VLLLALAVVAILGP-GVTTTILAIGIVYTPIFARVARASTLGVRTE
Mvan_3775|M.vanbaalenii_PYR-1 AFP-VLLLALAVVAILGP-GVTTTILAIAIVYTPIFARVARASTLGVRTE
TH_2705|M.thermoresistible__bu AFP-VLLLALAIVAILGP-GMTTTMLAIGIVFTPIFARVARAGTLGVRVE
MSMEG_6867|M.smegmatis_MC2_155 AFP-AILLALAVVAFLGN-GLWVVIIVLGVTNWVSYARVARSSVLSLRER
Mb3688c|M.bovis_AF2122/97 GLP-LLLAAIVLMQVMHHRTVWTVIAILALFGWPQVARIARGAVLEVRAS
Rv3664c|M.tuberculosis_H37Rv GLP-LLLAAIVLMQVMHHRTVWTVIAILALFGWPQVARIARGAVLEVRAS
MAV_0466|M.avium_104 GLP-LLLAAIVLMQVLHHRTVWTVIAILALFGWPQVARIARGAVLAVRAS
MMAR_5152|M.marinum_M GLP-LLLAAIVLMQVMHHRTVWTVIAILALFGWPQIARIARSSVLEVRGS
MUL_4238|M.ulcerans_Agy99 GLP-LLLAAIVLMQVMHHRTVWTVIAILALFGWPQIARIARSSVLEVRGS
MAB_0428|M.abscessus_ATCC_1997 GIP-TILGGMILLSVVGHRTVWSVAGALIAFGWMTSMRLVRSSVMSIKQS
MLBr_01123|M.leprae_Br4923 VVPSFVLIAIMSPQTKSSDNIMLLVLLLAGFGWMVSSRMVRGMTMSLRKS
.* :* .: : : *:.*. .: ::
Mflv_2759|M.gilvum_PYR-GCK PYVSVSRAMGAGDLYILRRHILPNIAGPLIVQTSLSLAFAILSEAALSFL
Mvan_3775|M.vanbaalenii_PYR-1 PYVSASRAMGTGDLYILRRHILPNIAGPLIVQTSLSLAFAILSEAALSFL
TH_2705|M.thermoresistible__bu PFVQASHTMGTGHAYILRRHILPNIAAPLIVQTSLSLAFAILAEAALSFL
MSMEG_6867|M.smegmatis_MC2_155 DYVLEARAIGVGPATIMRRHLLPNVLASLITIAALNVASAIVVESALSFL
Mb3688c|M.bovis_AF2122/97 DYVLAAKALGLNRFQILLRHALPNAVGPVIAVATVALGIFIVTEATLSYL
Rv3664c|M.tuberculosis_H37Rv DYVLAAKALGLNRFQILLRHALPNAVGPVIAVATVALGIFIVTEATLSYL
MAV_0466|M.avium_104 DYVLAAKALGMSRFQILIRHALPNALGPVIAVATIALGLFIVTEATLSYL
MMAR_5152|M.marinum_M DFVLAAKALGMSRFQILLRHALPNAIGPVIAVATVALGIFIVTEATLSYL
MUL_4238|M.ulcerans_Agy99 DFVLAAKALGMSRFQILLRHALPNAIGPVIAVATVALGIFIVTEATLSYL
MAB_0428|M.abscessus_ATCC_1997 DYVQAAIALGAPTWRILLRHILPNALAPVLVYATIAVGSFIAAEATLTYM
MLBr_01123|M.leprae_Br4923 EFIRAGKYMGVSSRRMIVGHVVPNVASILIIDATLNVGSAILAETGLSFL
:: . :* :: * :** . :: ::: :. * *: *:::
Mflv_2759|M.gilvum_PYR-GCK GLGIQPPQPSLGRMIFDSQGFVTMAWWMAVFPGAAIFLIVLAFNLFGDGL
Mvan_3775|M.vanbaalenii_PYR-1 GLGIQPPQPSLGRMIFDSQGFVTMAWWMAVFPGAAIFVIVLAFNLLGDGL
TH_2705|M.thermoresistible__bu GLGIQPPEPSLGRMIFDSQGFVTLAWWMAVFPGAAIVAAVLACNLFGDGL
MSMEG_6867|M.smegmatis_MC2_155 GLGVPADIPTWGSMLADGQLYLGTSWWIAVFPGLALMLTALSINITGDAI
Mb3688c|M.bovis_AF2122/97 GVGLPTSVVSWGGDINVAQTRLRSGSPILFYPAGALAITVLAFMMMGDAL
Rv3664c|M.tuberculosis_H37Rv GVGLPTSVVSWGGDINVAQTRLRSGSPILFYPAGALAITVLAFMMMGDAL
MAV_0466|M.avium_104 GVGLPPSVVSWGGDINLAQTRLRAGSPILFYPAGALAATVLAFMMMGDAL
MMAR_5152|M.marinum_M GVGLPPTVVSWGGDINVAQMRLRAGSPILFYPAGALAVTVLAFMMMGDAL
MUL_4238|M.ulcerans_Agy99 GVGLPPTVVSWGDDINVAQMRLRAGSPILFYPAGALAVTVLAFMMMGDAL
MAB_0428|M.abscessus_ATCC_1997 GIGLELPEISWGLQINDGQSRFASTPRLVIVPALFLSAAVLGFIMVGDAL
MLBr_01123|M.leprae_Br4923 GFGIKPPDVSLGTLIANGTQSATTFPWVFLFPAGVLVVILVCANLTGDGL
*.*: : * : . : . *. : : : **.:
Mflv_2759|M.gilvum_PYR-GCK RDVLDPKQRTTIEARRLQR-------------
Mvan_3775|M.vanbaalenii_PYR-1 RDVLDPKQRTTVEARRRER-------------
TH_2705|M.thermoresistible__bu RDVLDPKQRTVIETQRRARQRPGRGRRAGSAW
MSMEG_6867|M.smegmatis_MC2_155 RDAVDPRTYR----------------------
Mb3688c|M.bovis_AF2122/97 RDALDPASRAWRA-------------------
Rv3664c|M.tuberculosis_H37Rv RDALDPASRAWRA-------------------
MAV_0466|M.avium_104 RDALDPASRAWRA-------------------
MMAR_5152|M.marinum_M RDALDPASRAWRA-------------------
MUL_4238|M.ulcerans_Agy99 RDALDPASRAWRA-------------------
MAB_0428|M.abscessus_ATCC_1997 RDALDPRRR-----------------------
MLBr_01123|M.leprae_Br4923 RDAFDPVSKHLRR-------------------
**..**