For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNDTLIRLHRRLEQSATTDGLLDVAYTTVDTPVGPLLLAATEIGLVRVAFATEDHDAVLDTLAQRLSPRI LRAPQRLADAARQIDEYFAGARTQFDVALDHSLSHGFRQLVQRRLSQIPYGHTMSYKGVAEIVGNPNAVR AVGSACSTNPLPVVVPCHRVLRSDGSLGGYIGGLDAKTALLELERAA
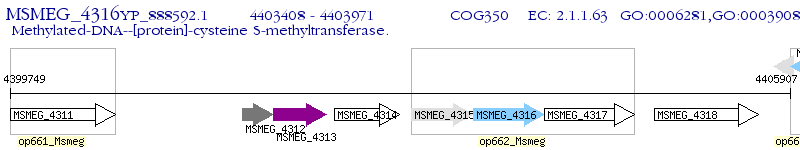
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4316 | - | - | 100% (187) | methylated-DNA--protein-cysteine methyltransferase |
| M. smegmatis MC2 155 | MSMEG_4928 | - | 8e-27 | 40.88% (159) | methylated-DNA--protein-cysteine methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1349c | ogt | 1e-26 | 40.83% (169) | methylated-DNA--protein-cysteine methyltransferase |
| M. gilvum PYR-GCK | Mflv_3909 | - | 4e-61 | 63.44% (186) | methylated-DNA--protein-cysteine methyltransferase |
| M. tuberculosis H37Rv | Rv1316c | ogt | 1e-26 | 40.83% (169) | methylated-DNA--protein-cysteine methyltransferase |
| M. leprae Br4923 | MLBr_01151 | ogt | 1e-26 | 43.29% (164) | putative methylated-DNA-protein-cysteine methyltransferase |
| M. abscessus ATCC 19977 | MAB_2332c | - | 2e-16 | 32.10% (162) | putative methylated-DNA:protein-cysteine methyltransferase |
| M. marinum M | MMAR_4082 | ogt | 1e-28 | 46.11% (167) | methylated-DNA--protein-cysteine methyltransferase Ogt |
| M. avium 104 | MAV_1536 | - | 6e-28 | 41.98% (162) | methylated-DNA--protein-cysteine methyltransferase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3946 | ogt | 2e-28 | 45.51% (167) | methylated-DNA--protein-cysteine methyltransferase Ogt |
| M. vanbaalenii PYR-1 | Mvan_2495 | - | 5e-60 | 63.74% (182) | methylated-DNA--protein-cysteine methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3909|M.gilvum_PYR-GCK MIDIAFDVPGFDIPTSDEDDTLRRLHDRLAEAAEPAGVLDVAYRTIDSPV
Mvan_2495|M.vanbaalenii_PYR-1 -----MSINMFDVTEADDADTLARLHGRLTAAADADGLLDVAYRTIDSPV
MSMEG_4316|M.smegmatis_MC2_155 -----------------MNDTLIRLHRRLEQSATTDGLLDVAYTTVDTPV
Mb1349c|M.bovis_AF2122/97 ---------------------------------------MIHYRTIDSPI
Rv1316c|M.tuberculosis_H37Rv ---------------------------------------MIHYRTIDSPI
MLBr_01151|M.leprae_Br4923 ---------------------------------------MIHCRSMDSPI
MAV_1536|M.avium_104 ---------------------------------------MIEYRTVDSPI
MMAR_4082|M.marinum_M ---------------------------------------MTHYRIVDSPI
MUL_3946|M.ulcerans_Agy99 ---------------------------------------MTHYRIIDSPI
MAB_2332c|M.abscessus_ATCC_199 ---------------------------------------MTDHATLTTPV
: :*:
Mflv_3909|M.gilvum_PYR-GCK GTLLLAATPAGLVRVAFDVEGHEVVLARLAESVSPRILRAPDRLDDVARQ
Mvan_2495|M.vanbaalenii_PYR-1 GSLLLAATPVGVVRVAFDVEIHDAVLARLAETLSPRILYAPHRLDAVARQ
MSMEG_4316|M.smegmatis_MC2_155 GPLLLAATEIGLVRVAFATEDHDAVLDTLAQRLSPRILRAPQRLADAARQ
Mb1349c|M.bovis_AF2122/97 GPLTLAGHGSVLTNLRMLEQTYEPSRTHWTP--------DPGAFSGAVDQ
Rv1316c|M.tuberculosis_H37Rv GPLTLAGHGSVLTNLRMLEQTYEPSRTHWTP--------DPGAFSGAVDQ
MLBr_01151|M.leprae_Br4923 GPLILAGHGSVLTNLQMVDQTYEPSRVGWLP--------ENGAFAEAVSQ
MAV_1536|M.avium_104 GLLTLAGRDPVLTNLRMVDQTYEPSRTGWTE--------NPRAFAGAVEQ
MMAR_4082|M.marinum_M GPLTLAGHDGVLTNLRMVDQTYEPSRVGWEV--------DDEAFADAVSQ
MUL_3946|M.ulcerans_Agy99 GPLTLAGHDGVLTNLRMVDQTYEPSRVGWEV--------DDEAFADAVSQ
MAB_2332c|M.abscessus_ATCC_199 GPFTAIVDDTGAVLASGWTDSPELLRALIHPSLRPKTLTPKRDLGKVTEA
* : . : : : ..
Mflv_3909|M.gilvum_PYR-GCK LDEYFAKRRTTFDVAVDLTLARGFRRNVVEHLRHIGYGRRASYADVAAAV
Mvan_2495|M.vanbaalenii_PYR-1 LDEYFAKQRTTFDVPVDLRLADGFRRSVVEHLRDIGYGRRESYAQVAAAI
MSMEG_4316|M.smegmatis_MC2_155 IDEYFAGARTQFDVALDHSLSHGFRQLVQRRLSQIPYGHTMSYKGVAEIV
Mb1349c|M.bovis_AF2122/97 LNAYFAGELTEFDVELDLRGTD-FQQRVWKALLTIPYGETRSYGEIADQI
Rv1316c|M.tuberculosis_H37Rv LNAYFAGELTEFDVELDLRGTD-FQQRVWKALLTIPYGETRSYGEIADQI
MLBr_01151|M.leprae_Br4923 LDAYFAGELTEFAIELDLCGTE-FQQRVWQALLTIPYGETRSYGEIAEQV
MAV_1536|M.avium_104 LGAYFAGELTEFDIELDLRGSE-FQRRVWRALQTIPYGETRSYGEIAEQI
MMAR_4082|M.marinum_M LDAYFAGERFDFDLALDLRGTE-FQRRVWRALLTIPYGETRSYGEIAERI
MUL_3946|M.ulcerans_Agy99 LDAYFAGERFDFDLALDLRGTE-FQRRVWQALLTIPYGETRSYGEIAERI
MAB_2332c|M.abscessus_ATCC_199 ITDYHAGDLTCIDSIPVVQRSGAFLMHAWDVLRTVDPQAPITYSEFADRA
: *.* : : * . * : :* .*
Mflv_3909|M.gilvum_PYR-GCK GSPRAVRAVGSACAHNPLPVVIPCHRVVRSDGTSGQYIGGPEAKTALLQL
Mvan_2495|M.vanbaalenii_PYR-1 GSPRAVRAVGTACAHNPLPVVIPCHRVVRSDGTIGQYVGGPHAKTALLDL
MSMEG_4316|M.smegmatis_MC2_155 GNPNAVRAVGSACSTNPLPVVVPCHRVLRSDGSLGGYIGGLDAKTALLEL
Mb1349c|M.bovis_AF2122/97 GAPGAARAVGLANGHNPIAIIVPCHRVIGASGKLTGYGGGINRKRALLEL
Rv1316c|M.tuberculosis_H37Rv GAPGAARAVGLANGHNPIAIIVPCHRVIGASGKLTGYGGGINRKRALLEL
MLBr_01151|M.leprae_Br4923 GAPGAARAVGLANSRNPIAIIVPCHRVIGASGQLIGYGGGLNRKLTLLEL
MAV_1536|M.avium_104 GAPGAARAVGLANGHNPIAIVVPCHRVIGASGKLTGYGGGLDRKQTLLAL
MMAR_4082|M.marinum_M GARGAARAVGLANGRNPVAVIVPCHRVIGASGGLTGYGGGLDRKQALLEL
MUL_3946|M.ulcerans_Agy99 GARGAARAVGLANGRNPVAVVVPCHRVIGASGGLTGYGGGLDRKQALLEL
MAB_2332c|M.abscessus_ATCC_199 GRPAAIRAAASACARNAAALFVPCHRVFRIGGALGGFRYGLPIKRWLLDH
* * **.. * . *. .:.:*****. .* : * * **
Mflv_3909|M.gilvum_PYR-GCK ESSSG---------------
Mvan_2495|M.vanbaalenii_PYR-1 EAVRE---------------
MSMEG_4316|M.smegmatis_MC2_155 ERAA----------------
Mb1349c|M.bovis_AF2122/97 EKSRAP------ADLTLFD-
Rv1316c|M.tuberculosis_H37Rv EKSRAP------ADLTLFD-
MLBr_01151|M.leprae_Br4923 EKHQVL------TSLTLFD-
MAV_1536|M.avium_104 ERRHSP------ASLTLFD-
MMAR_4082|M.marinum_M ERACVPADLTVRADLTLFDC
MUL_3946|M.ulcerans_Agy99 ERACVPADLTVRADLTLFDC
MAB_2332c|M.abscessus_ATCC_199 EAAAVR--------------
*