For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IHCRSMDSPIGPLILAGHGSVLTNLQMVDQTYEPSRVGWLPENGAFAEAVSQLDAYFAGELTEFAIELDL CGTEFQQRVWQALLTIPYGETRSYGEIAEQVGAPGAARAVGLANSRNPIAIIVPCHRVIGASGQLIGYGG GLNRKLTLLELEKHQVLTSLTLFD*
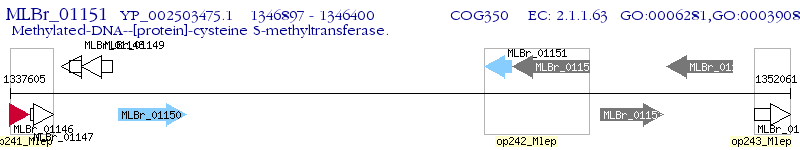
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01151 | ogt | - | 100% (165) | putative methylated-DNA-protein-cysteine methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1349c | ogt | 9e-73 | 78.18% (165) | methylated-DNA--protein-cysteine methyltransferase |
| M. gilvum PYR-GCK | Mflv_2325 | - | 8e-53 | 65.49% (142) | methylated-DNA--protein-cysteine methyltransferase |
| M. tuberculosis H37Rv | Rv1316c | ogt | 9e-73 | 78.18% (165) | methylated-DNA--protein-cysteine methyltransferase |
| M. abscessus ATCC 19977 | MAB_3012 | - | 3e-18 | 43.80% (121) | methylated-DNA--protein-cysteine methyltransferase |
| M. marinum M | MMAR_4082 | ogt | 3e-66 | 73.10% (171) | methylated-DNA--protein-cysteine methyltransferase Ogt |
| M. avium 104 | MAV_1536 | - | 1e-66 | 75.76% (165) | methylated-DNA--protein-cysteine methyltransferase |
| M. smegmatis MC2 155 | MSMEG_4928 | - | 7e-55 | 62.50% (160) | methylated-DNA--protein-cysteine methyltransferase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3946 | ogt | 1e-66 | 73.10% (171) | methylated-DNA--protein-cysteine methyltransferase Ogt |
| M. vanbaalenii PYR-1 | Mvan_4323 | - | 3e-56 | 64.60% (161) | methylated-DNA--protein-cysteine methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2325|M.gilvum_PYR-GCK ----MQYRVMDSPVGPLTLAGSHGRLQHLRMVDQTYEPDRADWMRDD---
Mvan_4323|M.vanbaalenii_PYR-1 -MDNMRYRTMDSPVGLLTLAGSTGRLQHLRMVDQTYEPDRAGWEKDD---
Mb1349c|M.bovis_AF2122/97 ---MIHYRTIDSPIGPLTLAGHGSVLTNLRMLEQTYEPSRTHWTPDP---
Rv1316c|M.tuberculosis_H37Rv ---MIHYRTIDSPIGPLTLAGHGSVLTNLRMLEQTYEPSRTHWTPDP---
MLBr_01151|M.leprae_Br4923 ---MIHCRSMDSPIGPLILAGHGSVLTNLQMVDQTYEPSRVGWLPEN---
MAV_1536|M.avium_104 ---MIEYRTVDSPIGLLTLAGRDPVLTNLRMVDQTYEPSRTGWTENP---
MMAR_4082|M.marinum_M ---MTHYRIVDSPIGPLTLAGHDGVLTNLRMVDQTYEPSRVGWEVDD---
MUL_3946|M.ulcerans_Agy99 ---MTHYRIIDSPIGPLTLAGHDGVLTNLRMVDQTYEPSRVGWEVDD---
MSMEG_4928|M.smegmatis_MC2_155 MRRALKCRTVDSPVGPLTLAGRDGHLVHLRMEDQTYEPSRDGWEVDD---
MAB_3012|M.abscessus_ATCC_1997 -MSTVGHCFFDTAIGSCAIAWSDRGIVALQLPERDDAATLARIRRPAGSE
.*:.:* :* : *:: :: .
Mflv_2325|M.gilvum_PYR-GCK ----TAFDAVIAQLQEYFAGERKDFALD-LELVGT-PFQRRVWEALLTIP
Mvan_4323|M.vanbaalenii_PYR-1 ----SAFPEAVMQLQEYFRGERREFDLD-LELAGT-AFQRRVWEALLTIP
Mb1349c|M.bovis_AF2122/97 ----GAFSGAVDQLNAYFAGELTEFDVE-LDLRGT-DFQQRVWKALLTIP
Rv1316c|M.tuberculosis_H37Rv ----GAFSGAVDQLNAYFAGELTEFDVE-LDLRGT-DFQQRVWKALLTIP
MLBr_01151|M.leprae_Br4923 ----GAFAEAVSQLDAYFAGELTEFAIE-LDLCGT-EFQQRVWQALLTIP
MAV_1536|M.avium_104 ----RAFAGAVEQLGAYFAGELTEFDIE-LDLRGS-EFQRRVWRALQTIP
MMAR_4082|M.marinum_M ----EAFADAVSQLDAYFAGERFDFDLA-LDLRGT-EFQRRVWRALLTIP
MUL_3946|M.ulcerans_Agy99 ----EAFADAVSQLDAYFAGERFDFDLA-LDLRGT-EFQRRVWQALLTIP
MSMEG_4928|M.smegmatis_MC2_155 ----SAFPEVVEQLAEYFAGERTDFELS-LDLVGT-AFQRTVWTALREIP
MAB_3012|M.abscessus_ATCC_1997 LAPPDFVAQAIDGIRRVLAGDDDDLQWARLDLDGTTDFDREVYAATRAIG
. .: : : *: :: *:* *: *:: *: * *
Mflv_2325|M.gilvum_PYR-GCK YGETRSYGEVARQIGAPGAFRAVGLANGRNPIGIIIPCHRVIGTNGSLTG
Mvan_4323|M.vanbaalenii_PYR-1 YGETRSYGQIAQQIDAPGAFRAVGLANGHNPIGIVVPCHRVIGANGSLTG
Mb1349c|M.bovis_AF2122/97 YGETRSYGEIADQIGAPGAARAVGLANGHNPIAIIVPCHRVIGASGKLTG
Rv1316c|M.tuberculosis_H37Rv YGETRSYGEIADQIGAPGAARAVGLANGHNPIAIIVPCHRVIGASGKLTG
MLBr_01151|M.leprae_Br4923 YGETRSYGEIAEQVGAPGAARAVGLANSRNPIAIIVPCHRVIGASGQLIG
MAV_1536|M.avium_104 YGETRSYGEIAEQIGAPGAARAVGLANGHNPIAIVVPCHRVIGASGKLTG
MMAR_4082|M.marinum_M YGETRSYGEIAERIGARGAARAVGLANGRNPVAVIVPCHRVIGASGGLTG
MUL_3946|M.ulcerans_Agy99 YGETRSYGEIAERIGARGAARAVGLANGRNPVAVVVPCHRVIGASGGLTG
MSMEG_4928|M.smegmatis_MC2_155 YGQTCSYGEIARKIGSPGASRAVGLANGHNPIGIIVPCHRVIGANGSLTG
MAB_3012|M.abscessus_ATCC_1997 PGRTRSYGEVAASVGEPGAAQAVGQSLGRNPIPLLVPCHRVLAADRSLHG
*.* ***::* :. ** :*** : .:**: :::*****:.:. * *
Mflv_2325|M.gilvum_PYR-GCK YG--GGLAQ--IFRGLLLVTAC------R-------
Mvan_4323|M.vanbaalenii_PYR-1 YG--GGLDRKKMLLGMERKNSP------TIPTLFD-
Mb1349c|M.bovis_AF2122/97 YG--GGINRKRALLELEKSRAP------ADLTLFD-
Rv1316c|M.tuberculosis_H37Rv YG--GGINRKRALLELEKSRAP------ADLTLFD-
MLBr_01151|M.leprae_Br4923 YG--GGLNRKLTLLELEKHQVL------TSLTLFD-
MAV_1536|M.avium_104 YG--GGLDRKQTLLALERRHSP------ASLTLFD-
MMAR_4082|M.marinum_M YG--GGLDRKQALLELERACVPADLTVRADLTLFDC
MUL_3946|M.ulcerans_Agy99 YG--GGLDRKQALLELERACVPADLTVRADLTLFDC
MSMEG_4928|M.smegmatis_MC2_155 YG--GGLERKRMLLDLEKSRMA--------PALF--
MAB_3012|M.abscessus_ATCC_1997 FSAYGGVVTKRELLRLEHTPGF------DDPTLF--
:. **: : :