For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TIAVTGSIATDHLMKFPGKFSEQLLADHLQKVSLSFLVDDLVIHRGGVAGNMAYAIGVLGGDVALVGAVG KDFDDYRGWLTTHGVNCDSVLVSESAYTARFVCTTDQDMAQIASFYPGAMSEARNISLAEVVATHGKPDL VIVGANDPDAMFGHTEECRKLGLAFAADPSQQLARLSGEEIRRLIDGATYLFTNDYEWDLLLQKSGWSEA EVMGQIELRVTTLGEKGVDLVGRDGTFVHVDVVPETHKEDPTGIGDAFRAGFLTGRSAGLSLERSAQLAS MVATLVLEAPGPQEWTWDRADAVRRLSEAYGAEAGTEIEQALSA*
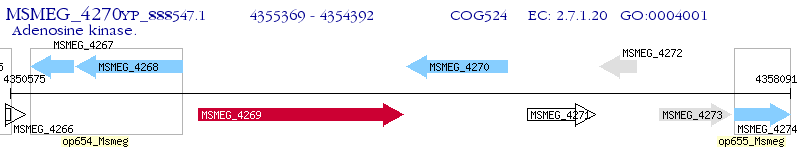
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4270 | - | - | 100% (325) | adenosine kinase |
| M. smegmatis MC2 155 | MSMEG_6801 | - | 2e-05 | 23.64% (275) | kinase, pfkB family protein |
| M. smegmatis MC2 155 | MSMEG_0504 | - | 2e-05 | 33.33% (120) | carbohydrate kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2225c | cbhK | 1e-145 | 78.09% (324) | carbohydrate kinase CbhK |
| M. gilvum PYR-GCK | Mflv_2947 | - | 1e-163 | 86.69% (323) | adenosine kinase |
| M. tuberculosis H37Rv | Rv2202c | cbhK | 1e-145 | 78.09% (324) | carbohydrate kinase CbhK |
| M. leprae Br4923 | MLBr_00873 | - | 1e-147 | 79.63% (324) | putative carbohydrate kinase |
| M. abscessus ATCC 19977 | MAB_1959 | - | 1e-136 | 73.37% (323) | adenosine kinase CbhK |
| M. marinum M | MMAR_3246 | cbhK | 1e-146 | 78.40% (324) | carbohydrate kinase CbhK |
| M. avium 104 | MAV_2289 | - | 1e-141 | 75.62% (324) | adenosine kinase |
| M. thermoresistible (build 8) | TH_2399 | cbhK | 1e-140 | 84.35% (294) | Probable carbohydrate kinase CbhK |
| M. ulcerans Agy99 | MUL_3559 | cbhK | 1e-145 | 78.09% (324) | carbohydrate kinase CbhK |
| M. vanbaalenii PYR-1 | Mvan_3566 | - | 1e-162 | 86.38% (323) | adenosine kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2947|M.gilvum_PYR-GCK MTIAVTGSIATDHLMRFPGKFSEQLLADHLQKVSLSFLVDDLVMHRGGVA
Mvan_3566|M.vanbaalenii_PYR-1 MTIAVTGSIATDHLMRFPGKFSEQLLADHLQKVSLSFLVDDLVMHRGGVA
MSMEG_4270|M.smegmatis_MC2_155 MTIAVTGSIATDHLMKFPGKFSEQLLADHLQKVSLSFLVDDLVIHRGGVA
Mb2225c|M.bovis_AF2122/97 MTIAVTGSIATDHLMRFPGRFSEQLLPEHLHKVSLSFLVDDLVMHRGGVA
Rv2202c|M.tuberculosis_H37Rv MTIAVTGSIATDHLMRFPGRFSEQLLPEHLHKVSLSFLVDDLVMHRGGVA
MMAR_3246|M.marinum_M MTIAVTGSIATDHLMRFPGHFSEQLLPEHLHKVSLSFLVDDLVVHRGGVA
MUL_3559|M.ulcerans_Agy99 MTIAVTGSIATDHLMRFPGHFSEQLLPEHLHKVSLSFLVDDLVVHRGGVA
MAV_2289|M.avium_104 MTIAVTGSIATDNLMRFPGRFSEHLLAEHLQKVSLSFLVDDLVIHRGGVA
MLBr_00873|M.leprae_Br4923 MTIAVTGSIATDHLMRFPGRFSEQLLVDNLHKVSLSFLVDDLVIHRGGVA
MAB_1959|M.abscessus_ATCC_1997 MSIIVTGSIATDHLMNFPGKFSEQLLVEHLQKVSLSFLVDDLEIRRGGVA
TH_2399|M.thermoresistible__bu VTIAVTGSIATDHLMRFPGKFSEQLLADHLQKVSLSFLVDDLVVHRGGVA
::* ********:**.***:***:** ::*:*********** ::*****
Mflv_2947|M.gilvum_PYR-GCK GNMAFAMGVLGGNPTLVGAVGEDFDDYRNWLSAHGVNCDGVLVSQNAYTA
Mvan_3566|M.vanbaalenii_PYR-1 GNMAFAMGVLGGSPTLVGAVGEDFDEYRQWLTRHGVNCDTVLVSRSAYTA
MSMEG_4270|M.smegmatis_MC2_155 GNMAYAIGVLGGDVALVGAVGKDFDDYRGWLTTHGVNCDSVLVSESAYTA
Mb2225c|M.bovis_AF2122/97 GNMAFAIGVLGGEVALVGAAGADFADYRDWLKARGVNCDHVLISETAHTA
Rv2202c|M.tuberculosis_H37Rv GNMAFAIGVLGGEVALVGAAGADFADYRDWLKARGVNCDHVLISETAHTA
MMAR_3246|M.marinum_M GNMAFAIGVLGGDVALVGAAGADFADYRDWLKAHGVNCDHVLISQTAHTA
MUL_3559|M.ulcerans_Agy99 GNMAFAIGVLGGDVALVGAAGADFADYRDWLKAHGVNCDHVLISQTAHTA
MAV_2289|M.avium_104 GNIAFAIGVLGGDVALVGAAGDDFGEYRDWLQRHGVNCDHVLISQTAHTA
MLBr_00873|M.leprae_Br4923 GNMAYAIGVLGGDVALIGAVGDDFADYRDWLQNHGVNCDSVLISKTAHTA
MAB_1959|M.abscessus_ATCC_1997 GNIGFAIGVLGGKPTLVGAVGADFAEYRSWLEDHGVNCDAVLISETAHTA
TH_2399|M.thermoresistible__bu GNMAYAIGRLGGSVALVGAAGPDFAEYRQWLESVNVDCGCVLLSETAYTA
**:.:*:* ***. :*:**.* ** :** ** .*:*. **:*..*:**
Mflv_2947|M.gilvum_PYR-GCK RFVCTTDEDMAQIASFYPGAMSEARNIKLADIVTRTGTPDLVIIGANDPE
Mvan_3566|M.vanbaalenii_PYR-1 RFVCTTDEDMAQIASFYPGAMSEARNIKLADIVKQTGTPDLVIIGANDPE
MSMEG_4270|M.smegmatis_MC2_155 RFVCTTDQDMAQIASFYPGAMSEARNISLAEVVATHGKPDLVIVGANDPD
Mb2225c|M.bovis_AF2122/97 RFTCTTDVDMAQIASFYPGAMSEARNIKLADVVSAIGKPELVIIGANDPE
Rv2202c|M.tuberculosis_H37Rv RFTCTTDVDMAQIASFYPGAMSEARNIKLADVVSAIGKPELVIIGANDPE
MMAR_3246|M.marinum_M RFTCTTDVDMAQIASFYPGAMSEARNIKLADVVSAVGTPELVIVGANDPE
MUL_3559|M.ulcerans_Agy99 RFTCTTDVDMAQIASFYPGAMSEARNIKLADVVSAVGTPELVIVAANDPE
MAV_2289|M.avium_104 RFVCTTDQDMAQIASFYPGAMSEARNIKLADLVSSLGKPELVIIGANDPE
MLBr_00873|M.leprae_Br4923 RFVCTTDLDMAQIGSFYTGAMSEARNIKLVDAIATIGTPELVIIGANDPA
MAB_1959|M.abscessus_ATCC_1997 RFVCTTDQDMAQIASFYPGAMSEARDISLADVVSRTGTPELVIIGANDPD
TH_2399|M.thermoresistible__bu RFVCTSDEDMAQIASFYPGAMSEARNISLADVVARIGTPELVIIGANDPE
**.**:* *****.***.*******:*.*.: : *.*:***:.****
Mflv_2947|M.gilvum_PYR-GCK AMFLYTEECRALGLPFAADPSQQLARLSGDEIRKLIDGATYLFTNDYEWD
Mvan_3566|M.vanbaalenii_PYR-1 AMFLYTEECRTLGLAFAADPSQQLARLSGEEIRRLIDGAAYLFTNDYEWD
MSMEG_4270|M.smegmatis_MC2_155 AMFGHTEECRKLGLAFAADPSQQLARLSGEEIRRLIDGATYLFTNDYEWD
Mb2225c|M.bovis_AF2122/97 AMFLHTEECRKLGLAFAADPSQQLARLSGEEIRRLVNGAAYLFTNDYEWD
Rv2202c|M.tuberculosis_H37Rv AMFLHTEECRKLGLAFAADPSQQLARLSGEEIRRLVNGAAYLFTNDYEWD
MMAR_3246|M.marinum_M AMFLHTEECRQLGLAFAADPSQQLARLSGDEIKRLINGATYLFTNDYEWD
MUL_3559|M.ulcerans_Agy99 AMFLHTEECRQLGLAFAADPSQQLARLSGDEIKRLINGATYLFTNDYEWD
MAV_2289|M.avium_104 AMVVHTEECRKLGLPFAADPSQQLPRLSGEEIDKLVDGAAYLFTNDYEWE
MLBr_00873|M.leprae_Br4923 AMLGYTQECRKLGLAFAADPSQQLARLSGEEIRNLIDGATYLFTNDYEWD
MAB_1959|M.abscessus_ATCC_1997 AMFRHTEECRSLGLPFAADPSQQLARLSGEQARDLVGGAAYLFTNDYEWD
TH_2399|M.thermoresistible__bu AMFRHTEECRELGLAFAADPSQQLARLSGEEIRRLINGATYLFTNDYEWD
**. :*:*** ***.*********.****:: *:.**:*********:
Mflv_2947|M.gilvum_PYR-GCK LLLQKSGWSEAEVMNQIQLRVTTLGEKGVDLVGRDGTFVHVDVVPEIHKE
Mvan_3566|M.vanbaalenii_PYR-1 LLLQKSGWSEAEVMSQIQLRVTTLGEKGVDLVGRDGTFVHVDVVPETHKE
MSMEG_4270|M.smegmatis_MC2_155 LLLQKSGWSEAEVMGQIELRVTTLGEKGVDLVGRDGTFVHVDVVPETHKE
Mb2225c|M.bovis_AF2122/97 LLLSKTGWSEADVMAQIDLRVTTLGPKGVDLVEPDGTTIHVGVVPETSQT
Rv2202c|M.tuberculosis_H37Rv LLLSKTGWSEADVMAQIDLRVTTLGPKGVDLVEPDGTTIHVGVVPETSQT
MMAR_3246|M.marinum_M LLLSKTGWSEADVMAQIGLRVTTLGPKGVDLVEPDGTSIHVGVVPETSQT
MUL_3559|M.ulcerans_Agy99 LLLSKTGWSEADVMAQIGLRVTTLGPKGVDLVEPDGTSIHVGVVPETSQT
MAV_2289|M.avium_104 LLLSKTGWSEADVQQKVGLRVTTLGAKGVDIVERDGTTTRVGVVPETSQT
MLBr_00873|M.leprae_Br4923 LLLSKTGWSEADVMAQIQLRVTTLGAKGVDLVEPDGTRVHVSVVPEAGQV
MAB_1959|M.abscessus_ATCC_1997 LLLSKTGWTEADVRERVELRITTLGANGVEIVEKDGTSLKVGVVPEKGQV
TH_2399|M.thermoresistible__bu LLLTKTGWSEAQVMSQIGLRVTTLGPKGVDLVSSDGTFVHVGVVPEKERV
*** *:**:**:* :: **:**** :**::* *** :*.**** :
Mflv_2947|M.gilvum_PYR-GCK DPTGIGDAFRAGFLTGRDAGLSLERSAQLASLVATLVLEAPGPQE-----
Mvan_3566|M.vanbaalenii_PYR-1 DPTGIGDAFRAGFLTGRDAGLSLERSAQLASMVATLVLEAPGPQE-----
MSMEG_4270|M.smegmatis_MC2_155 DPTGIGDAFRAGFLTGRSAGLSLERSAQLASMVATLVLEAPGPQE-----
Mb2225c|M.bovis_AF2122/97 DPTGVGDAFRAGFLTGRSAGLGLERSAQLGSLVAVLVLESTGTQE-----
Rv2202c|M.tuberculosis_H37Rv DPTGVGDAFRAGFLTGRSAGLGLERSAQLGSLVAVLVLESTGTQE-----
MMAR_3246|M.marinum_M DPTGVGDAFRAGFLTGRSAGLNLERAAQLGSLVAVLVLESTGTQE-----
MUL_3559|M.ulcerans_Agy99 DPTGVGDAFRAGFLTGRSAGLNLERAAQLGSLVAVLVLESTGTQE-----
MAV_2289|M.avium_104 DPTGVGDAFRAGFLTGRSAGLNLERSAQLGSLVAVLVLESTGTQE-----
MLBr_00873|M.leprae_Br4923 DPTGVGDAFRAGFLTGRSAGLSLERSAQLGSLVAVLVLESTGTQQ-----
MAB_1959|M.abscessus_ATCC_1997 DPTGVGDAWRAGFLSARSAGLNLERSAQLGSLVATLVLETVGTQE-----
TH_2399|M.thermoresistible__bu DPTGIGDAFRAGFLTGRSAGLSLERSAQLASLVAVLVLEAPGPRXXLRPV
****:***:*****:.*.***.***:***.*:**.****: *.:
Mflv_2947|M.gilvum_PYR-GCK -----------------------WHWDKD-------AAVRRLSEAYGADA
Mvan_3566|M.vanbaalenii_PYR-1 -----------------------WTWNKD-------SAVQRLTDAYGADA
MSMEG_4270|M.smegmatis_MC2_155 -----------------------WTWDRA-------DAVRRLSEAYGAEA
Mb2225c|M.bovis_AF2122/97 -----------------------WQWDYE-------AAASRLAGAYGEHA
Rv2202c|M.tuberculosis_H37Rv -----------------------WQWDYE-------AAASRLAGAYGEHA
MMAR_3246|M.marinum_M -----------------------WQWDRQ-------VATARLAEAYGDDA
MUL_3559|M.ulcerans_Agy99 -----------------------WQWDRQ-------VATARLAEAYGDDA
MAV_2289|M.avium_104 -----------------------WSWDHE-------VAKSRLAGAYGEDA
MLBr_00873|M.leprae_Br4923 -----------------------WGWDRD-------VAVARLAGAYGEEA
MAB_1959|M.abscessus_ATCC_1997 -----------------------WLWERD-------EALVRLADAYGQEA
TH_2399|M.thermoresistible__bu VPVAVVMCLPARRSDPVVPYRTTGRVDRNPVSRCATRSPGRHTAGRGRSG
: : * : . * .
Mflv_2947|M.gilvum_PYR-GCK GNEIAEAL-------
Mvan_3566|M.vanbaalenii_PYR-1 AADIAAAI-------
MSMEG_4270|M.smegmatis_MC2_155 GTEIEQALSA-----
Mb2225c|M.bovis_AF2122/97 AAEIVAVLA------
Rv2202c|M.tuberculosis_H37Rv AAEIVAVLA------
MMAR_3246|M.marinum_M ANEITAVLA------
MUL_3559|M.ulcerans_Agy99 ANEITAVLA------
MAV_2289|M.avium_104 AAEIAAVLA------
MLBr_00873|M.leprae_Br4923 AAEITVVLS------
MAB_1959|M.abscessus_ATCC_1997 ADEIGAAL-------
TH_2399|M.thermoresistible__bu AGPSPPSLRLRWRPR
. :