For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSQDNSVVPVIDIAALHGDDEAAMIAVAAELDAACREIGFFQIRNHGISEDVIEAMYRTADEFFALPDEE KRLVAQPSSDAVRGYSSIGEQAFSYSEDVHQPRDLHEKFDIGPVDVDRDDPYYAPENAGPHFLPNLWPQR PAGMEAAWTTYFHAMNDLARKLMSAFALGLRLPADYFVDTIDRDISMLRAINYPHLNTPPQPGQMRAGAH TDYGSLTIVRQEAAPGGLEVFTKDGDWISVPVVPDALVVNIGDLMAQWTNDLWTSTRHRVRTPGPDASGD TRRMSLVFFHQPNYDAVIETLPTCITADNPRRYDPTTSGDHLTSKFEKTIALASTNG
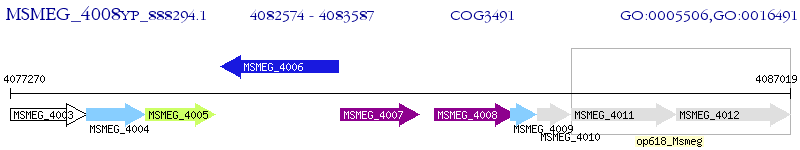
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4008 | - | - | 100% (337) | oxidoreductase, 2OG-Fe(II) oxygenase family protein |
| M. smegmatis MC2 155 | MSMEG_4007 | - | 2e-59 | 37.58% (306) | oxidoreductase, 2OG-Fe(II) oxygenase family protein |
| M. smegmatis MC2 155 | MSMEG_4685 | - | 5e-50 | 36.45% (299) | oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2580 | - | 1e-39 | 31.48% (324) | 2OG-Fe(II) oxygenase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4434c | - | 2e-27 | 29.10% (299) | putative iron/ascorbate dependent oxidoreductase |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_4913 | - | 9e-29 | 31.48% (324) | oxidoreductase, 2OG-Fe(II) oxygenase family protein |
| M. thermoresistible (build 8) | TH_4524 | - | 4e-42 | 38.12% (320) | PUTATIVE 2OG-Fe(II) oxygenase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4054 | - | 1e-46 | 33.64% (324) | 2OG-Fe(II) oxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2580|M.gilvum_PYR-GCK ----------MSVIATVDLSRWREG-GPAAEDVAAQVDAGLQRAGFILVT
Mvan_4054|M.vanbaalenii_PYR-1 ----------MSVIATVDLSRWRAG-GAAADAVAADVDEGLQRAGFILVS
MSMEG_4008|M.smegmatis_MC2_155 ------MSQDNSVVPVIDIAALHGDDEAAMIAVAAELDAACREIGFFQIR
TH_4524|M.thermoresistible__bu ------MAGDFTSVPVIDISALRHGSGPGYRRAVTALGDAARHVGFAQLV
MAB_4434c|M.abscessus_ATCC_199 MAVKTVAPKRLASNHGRVAELPVIDLSSDSNQLRAALREVAHEVGFFYLT
MAV_4913|M.avium_104 ------MSNDMCPVPG-VTAVPGVDLRADPDVLRDGLREAAHRVGFFYLT
. . : :. ** :
Mflv_2580|M.gilvum_PYR-GCK GHGVDPDLARDVRAAAREFFALDPTVKAGYSVPVG--GRGWIGPGAEANG
Mvan_4054|M.vanbaalenii_PYR-1 GHGVSASLARDVRAAAREFFALPDEVKSGYSVPVG--GHGWIGPGAEANG
MSMEG_4008|M.smegmatis_MC2_155 NHGISEDVIEAMYRTADEFFALPDEEKRLVAQPSSDAVRGYSSIGEQAFS
TH_4524|M.thermoresistible__bu GHGIDPALFDALVDRTRAFFALPAAEKHRVYIGNSTNHRGYVPPGEEVFA
MAB_4434c|M.abscessus_ATCC_199 GHGVSPQLVHEVLSAARKLFALSEADKEAIAMVRSPHFRGFTRLGGELTG
MAV_4913|M.avium_104 GHGVPADLVDRVLDAARRFFRLPQADKDAVAMVRSPHFRGYTRLGGERTR
.**: : : : :* * * . :*: * :
Mflv_2580|M.gilvum_PYR-GCK YAEGTETPPDLKESFSLG-----AETATGDAEVDRIWFAPNVWPREVPEL
Mvan_4054|M.vanbaalenii_PYR-1 YAEGTETPPDLKETFSLG-----AETATGDAEVDRIWFAPNVWPREVPAL
MSMEG_4008|M.smegmatis_MC2_155 YSEDVHQPRDLHEKFDIGPVDVDRDDPYYAPENAGPHFLPNLWPQRPAGM
TH_4524|M.thermoresistible__bu G-----GTADTKEAFDLS-YELPADDPRYLAG--DPLLGPNQWP-ELTGF
MAB_4434c|M.abscessus_ATCC_199 G------LTDWREQIDVG--PERAASPTADGRDYMWLQGPNQWPAAVPEL
MAV_4913|M.avium_104 G------AVDWREQIDIG--PQRPPIGGLDKSDYRWLQGPNQWPAALPEL
* :* :.:. ** ** . :
Mflv_2580|M.gilvum_PYR-GCK APLVEEYTAQMRRVADDLLALCAHALGLSVNPFAALAS-APTWTMNINHY
Mvan_4054|M.vanbaalenii_PYR-1 QPLLTEYTAQMRGVADDLLALFAHALKLPANPFAELAS-TPTWTMNINHY
MSMEG_4008|M.smegmatis_MC2_155 EAAWTTYFHAMNDLARKLMSAFALGLRLPADYFVDTID-RDISMLRAINY
TH_4524|M.thermoresistible__bu REAVTAWYDAVFALSRAILRAFAVALGQPPDRFDRYVT-TPPSQLRLIHY
MAB_4434c|M.abscessus_ATCC_199 RPALESWDIALAEVGRKLLREWAIALGSDGAVFSEAFGDKPATLIKVIRY
MAV_4913|M.avium_104 PGLIAEWDAALSGVARTLLRHWAAALGSPADVFDAAFADAPATLIKIIRY
: : :. :: * .* * :. .*
Mflv_2580|M.gilvum_PYR-GCK PPVSVVGEPEPGQFRIGPHTDFGTVTILDREPGAGGLQVYSEQ---AGWE
Mvan_4054|M.vanbaalenii_PYR-1 PPVSVVGEPEPGQFRIGPHTDFGTVTILDREPGAGGLQVYSED---AGWQ
MSMEG_4008|M.smegmatis_MC2_155 PHL--NTPPQPGQMRAGAHTDYGSLTIVRQEAAPGGLEVFTKD---GDWI
TH_4524|M.thermoresistible__bu P----ADPSAVDRPGIGAHTDYECFTLLRPTAP--GLEVLNAA---GQWI
MAB_4434c|M.abscessus_ATCC_199 P------SDGTTSQGVGAHRDSGVLTLLLTEPEVGGLQVEVDG----AWL
MAV_4913|M.avium_104 P------GRAAGPQGVGAHRDAGVLTLLLAEPGSRGLQVRRGGPRGDGWI
* *.* * .*:: . **:* *
Mflv_2580|M.gilvum_PYR-GCK NAPYDPAALTVNIGDLLEYWSGRRWPSGRHRVLPPQPD-APEEDLVSLIY
Mvan_4054|M.vanbaalenii_PYR-1 DAPYDPDALTVNIGDLLEYWSGRRWPSGRHRVLPPQPG-APEEDLVSLIY
MSMEG_4008|M.smegmatis_MC2_155 SVPVVPDALVVNIGDLMAQWTNDLWTSTRHRVRTPGPDASGDTRRMSLVF
TH_4524|M.thermoresistible__bu DVPYLDDAFVLNIGDLLEIWTNGAFVATTHRVRR------VTSERYSFPL
MAB_4434c|M.abscessus_ATCC_199 EAPGREGAFIVNIGELLERASGGYLRATRHRVTLTG------APRISVAY
MAV_4913|M.avium_104 DVPPRQGAFIVNIGELLEAATRGYLRATEHRVNLAGST----AERISVPY
..* *: :***:*: : : *** *.
Mflv_2580|M.gilvum_PYR-GCK FYEANHDALVT--PLSPPIGRAEGLEPVLSADFIKQRLDAITVS------
Mvan_4054|M.vanbaalenii_PYR-1 FYEANHDALVT--PLDPPVGRVSGLAPVTSADFIKERLDAITVG------
MSMEG_4008|M.smegmatis_MC2_155 FHQPNYDAVIETLPTCITADNPRRYDPTTSGDHLTSKFEKTIALASTNG-
TH_4524|M.thermoresistible__bu FVN--VDADTEVAPLAEFVDGTARYAPVIAGEHLFAQTARSFRYLQDRIR
MAB_4434c|M.abscessus_ATCC_199 FFNPRLDARIPVLELPPELRDRARG---VSADPDDPIHATYGENAWKSRL
MAV_4913|M.avium_104 FFNPRLDARIPVLSLPPELRARAGERWTPSEDPADPIFSVYGRNAWKSRL
* : ** : :
Mflv_2580|M.gilvum_PYR-GCK ------------------------
Mvan_4054|M.vanbaalenii_PYR-1 ------------------------
MSMEG_4008|M.smegmatis_MC2_155 ------------------------
TH_4524|M.thermoresistible__bu RGEVSLPDGAYPLSAFGRRAASPR
MAB_4434c|M.abscessus_ATCC_199 RAHPDVAAVHGHLDS---------
MAV_4913|M.avium_104 RSHPDVASAHGFSAHHGAAD----