For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RSDDSDPNVRKTARIGMIVPSSNTCLEPLTYRMLDNRCDVTVHFTRTPVTRVGLDDTSDAQFDTGAMQAA ATLLATADVDLIAWNGTAGSWLGTSHDRAITDAISRATGVRAVTSTLAFMAVFGALGVHRLGIVTPYPDY MNARIGESYRAEGVTVISTQGFGLTETTDIARIQPHELGSPTRHVAAAAPDAIAYVCTNLRGADAVEALE AELGVPVLDSVAVTLAVCLAMVGAAPLDARWGSRLSHGAGLAGETLART*
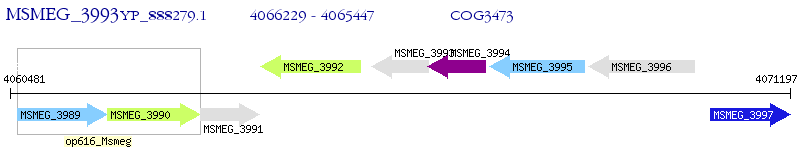
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3993 | - | - | 100% (260) | Asp/Glu racemase |
| M. smegmatis MC2 155 | MSMEG_1319 | - | 1e-46 | 45.22% (230) | Asp/Glu racemase |
| M. smegmatis MC2 155 | MSMEG_2490 | - | 4e-13 | 25.77% (194) | decarboxylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4413c | - | 1e-18 | 28.87% (239) | putative decarboxylase |
| M. marinum M | MMAR_3401 | - | 5e-15 | 25.40% (248) | maleate cis-trans isomerase |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3993|M.smegmatis_MC2_155 --MRSDDSDPNVRKTARIGMIVPSSNTCLEPLTYRMLDNR-CDVTVHFTR
MAB_4413c|M.abscessus_ATCC_199 MDFSSLPDFAESADQRGIGIIAP-FDLALERELWRWVP---AEVSLHLAR
MMAR_3401|M.marinum_M ------------MSSYRVGLLVPSSNTTMEAEVPEVLRANGSTTGDRFTF
. :*::.* : :* . : . . :::
MSMEG_3993|M.smegmatis_MC2_155 TPVTRVGLD-DTSDAQFDTGAMQAAATLLATADVDLIAWNGTAGSWLG-T
MAB_4413c|M.abscessus_ATCC_199 TPYEPVPVSRAMAELVSNTSHLQAATRDVLHVEPEVVAYLCTSGSFIRGV
MMAR_3401|M.marinum_M HASRMRMKKVDPQELMAMNASAERATTELADAAVDVMAYACLVAVMAAGP
. . : .. : *: : . :::*: .
MSMEG_3993|M.smegmatis_MC2_155 SHDRAITDAISRATGVR-----AVTSTLAFMAVFGALGVHRLGIVTPYPD
MAB_4413c|M.abscessus_ATCC_199 QHERELVDAIMAAGAKS-----AVTTSGALAEAVTALNINRISVITPYDA
MMAR_3401|M.marinum_M NAHRTIEERLTKVASQSGGTVEVISSAGALVEGIRELGAKKIAMVAPYMK
. .* : : : . . .:::: *: . *. :::.:::**
MSMEG_3993|M.smegmatis_MC2_155 YMNARIGESYRAEGVTVISTQGFGLTETTDIARIQPHELGSPTRHVAAAA
MAB_4413c|M.abscessus_ATCC_199 ELTELLAKFLGELGVTVVRTDHLGLGG--GIWKVNYRTVAERILAADTAD
MMAR_3401|M.marinum_M PLTSQVVDYIEAEGIEVMDVISLEVEDNRAVGRLDPMRLPHIAHRLDTHD
:. : . *: *: . : : : ::: : :
MSMEG_3993|M.smegmatis_MC2_155 PDAIAY-VCTNLRGADAVEALEAELGVPVLDSVAVTLAVCLAMVGAAPLD
MAB_4413c|M.abscessus_ATCC_199 AEAIFV-SCTNLRTYDVIDPLEQLLGKPVLTANQLTIWACLDRMSLPMMG
MMAR_3401|M.marinum_M ADAVVLSACVQMPSLAAIPAAEAMLGLPVISAATATSWKILRSLGVAPVA
.:*: *.:: .: . * ** **: : * * :. . :
MSMEG_3993|M.smegmatis_MC2_155 ARWGSRLSHGAGLAGETLART
MAB_4413c|M.abscessus_ATCC_199 PGRWLRDVFGGGGSSD-----
MMAR_3401|M.marinum_M ADAG--ALFDGGASG------
. ...* :.