For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRIGMIVPSSNTALEPATTRLLTDRPDVTVHYTRIPVRAITLDGGGAAFDADTMVAAARLLADAEVDCIV WNGTAGSWLGLEHDRRCCDAITEATGIPATTSTLAILRACVDFGISALACATPYTRDVVDAISREYARHD VGIVSHAEWELTDNLSFAKASSAEVAELLAAAAKNGDPAPQGVALICTNVDGTTVLREVEQSLGIPVIDS IAATLWWALELTGSDARIPAAGLLLEQGPLRHRVKEIVTELRVRTGCDRTTARIDDADLGLHVDLCAAES CAPGVRSIQHDPSLDQRKLETVSWLEQHRKVLVQPAFTEAPYPPQALREVYGVSAQVLGPVERGDEMAAW LSAHSIGERAWTGDDLAAMADAQEKLHALLNNK*
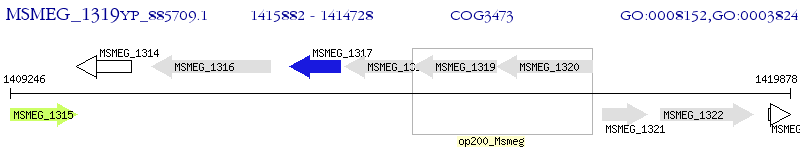
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1319 | - | - | 100% (384) | Asp/Glu racemase |
| M. smegmatis MC2 155 | MSMEG_3993 | - | 2e-46 | 45.22% (230) | Asp/Glu racemase |
| M. smegmatis MC2 155 | MSMEG_2490 | - | 6e-13 | 27.84% (194) | decarboxylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4413c | - | 1e-12 | 27.43% (226) | putative decarboxylase |
| M. marinum M | MMAR_3401 | - | 2e-08 | 22.98% (248) | maleate cis-trans isomerase |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1319|M.smegmatis_MC2_155 --MTRIGMIVPSSN----TALEPATTRLLTDR-----PDVTVHYTRIPVR
MMAR_3401|M.marinum_M MSSYRVGLLVPSSN----TTMEAEVPEVLRANGSTTGDRFTFHASRMRMK
MAB_4413c|M.abscessus_ATCC_199 MDFSSLPDFAESADQRGIGIIAPFDLALERELWRWVPAEVSLHLARTPYE
: :. *:: : . : .:.* :* .
MSMEG_1319|M.smegmatis_MC2_155 AIT---LDGGGAAFDADTMVAAARLLADAEVDCIVWNGTAGSWLGLEHDR
MMAR_3401|M.marinum_M KVDPQELMAMNASAERATTELADAAVDVMAYACLVAVMAAGPNAHRTIEE
MAB_4413c|M.abscessus_ATCC_199 PVP-VSRAMAELVSNTSHLQAATRDVLHVEPEVVAYLCTSGSFIRGVQHE
: : * : :. ::*. ..
MSMEG_1319|M.smegmatis_MC2_155 RCCDAITEATG-IPATTSTLAILRACVDFGISALACATPYTRDVVDAISR
MMAR_3401|M.marinum_M RLTKVASQSGGTVEVISSAGALVEGIRELGAKKIAMVAPYMKPLTSQVVD
MAB_4413c|M.abscessus_ATCC_199 RELVDAIMAAGAKSAVTTSGALAEAVTALNINRISVITPYDAELTELLAK
* : * . ::: *: .. :. . :: :** :.. :
MSMEG_1319|M.smegmatis_MC2_155 EYARHDVGIVSHAEWELTDNLSFAKASSAEVAELLAAAAKNGDPAPQGVA
MMAR_3401|M.marinum_M YIEAEGIEVMDVISLEVEDNRAVGRLDPMRLPHIAHRLDTHD--ADAVVL
MAB_4413c|M.abscessus_ATCC_199 FLGELGVTVVRTDHLGLGG--GIWKVNYRTVAERILAADTAD---AEAIF
.: :: : . .. : . :.. . . :
MSMEG_1319|M.smegmatis_MC2_155 LICTNVDGTTVLREVEQSLGIPVIDSIAATLWWALELTGSDARIPAAGL-
MMAR_3401|M.marinum_M SACVQMPSLAAIPAAEAMLGLPVISAATATSWKILRSLGVAPVAADAGA-
MAB_4413c|M.abscessus_ATCC_199 VSCTNLRTYDVIDPLEQLLGKPVLTANQLTIWACLDRMSLPMMGPGRWLR
*.:: .: * ** **: : * * * . .
MSMEG_1319|M.smegmatis_MC2_155 -LLEQGPLRHRVKEIVTELRVRTGCDRTTARIDDADLGLHVDLCAAESCA
MMAR_3401|M.marinum_M -LFDGGASG-----------------------------------------
MAB_4413c|M.abscessus_ATCC_199 DVFGGGGSSD----------------------------------------
:: *
MSMEG_1319|M.smegmatis_MC2_155 PGVRSIQHDPSLDQRKLETVSWLEQHRKVLVQPAFTEAPYPPQALREVYG
MMAR_3401|M.marinum_M --------------------------------------------------
MAB_4413c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_1319|M.smegmatis_MC2_155 VSAQVLGPVERGDEMAAWLSAHSIGERAWTGDDLAAMADAQEKLHALLNN
MMAR_3401|M.marinum_M --------------------------------------------------
MAB_4413c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_1319|M.smegmatis_MC2_155 K
MMAR_3401|M.marinum_M -
MAB_4413c|M.abscessus_ATCC_199 -