For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DFSSLPDFAESADQRGIGIIAPFDLALERELWRWVPAEVSLHLARTPYEPVPVSRAMAELVSNTSHLQAA TRDVLHVEPEVVAYLCTSGSFIRGVQHERELVDAIMAAGAKSAVTTSGALAEAVTALNINRISVITPYDA ELTELLAKFLGELGVTVVRTDHLGLGGGIWKVNYRTVAERILAADTADAEAIFVSCTNLRTYDVIDPLEQ LLGKPVLTANQLTIWACLDRMSLPMMGPGRWLRDVFGGGGSSD*
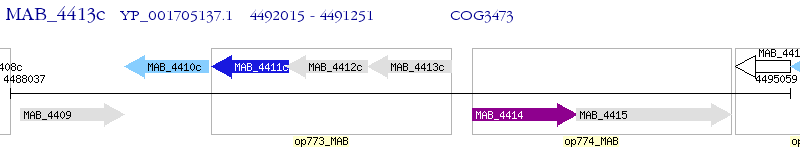
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4413c | - | - | 100% (254) | putative decarboxylase |
| M. abscessus ATCC 19977 | MAB_4412c | - | 3e-17 | 32.67% (150) | putative decarboxylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_3401 | - | 4e-11 | 24.02% (204) | maleate cis-trans isomerase |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_2495 | - | 1e-61 | 51.74% (230) | decarboxylase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4413c|M.abscessus_ATCC_199 MDFSSLPDFAESADQRGIGIIAP-FDLALERELWRWVPAEVSLHLARTPY
MSMEG_2495|M.smegmatis_MC2_155 ------------MEQTGIGVVVP-YDFALDRELWRWVPEHISLHLTRTPH
MMAR_3401|M.marinum_M ------------MSSYRVGLLVPSSNTTMEAEVPEVLRANGSTTGDRFTF
.. :*::.* : ::: *: . : . * * ..
MAB_4413c|M.abscessus_ATCC_199 EPVPVSRAMA---ELVSNTSHLQAATRDVLHVEPEVVAYLCTSGSFIRGV
MSMEG_2495|M.smegmatis_MC2_155 VPLSVTMEMA---IYVSNPNLLAQASASLHAVAPVVTAYACTSGSFVSGV
MMAR_3401|M.marinum_M HASRMRMKKVDPQELMAMNASAERATTELADAAVDVMAYACLVAVMAAGP
. : . :: *: .: . * ** * . : *
MAB_4413c|M.abscessus_ATCC_199 QHERELVDAIMAAGAKS-----AVTTSGALAEAVTALNINRISVITPYDA
MSMEG_2495|M.smegmatis_MC2_155 TGETEIVAAMTQAGAPA-----ALTTSGALLTALRHLDVSSIAAVTPYTA
MMAR_3401|M.marinum_M NAHRTIEERLTKVASQSGGTVEVISSAGALVEGIRELGAKKIAMVAPYMK
. : : ..: : .::::*** .: *. . *: ::**
MAB_4413c|M.abscessus_ATCC_199 ELTELLAKFLGELGVTVVRTDHLGLGG--GIWKVNYRTVAERILAADTAD
MSMEG_2495|M.smegmatis_MC2_155 DLTSGLVSFLGEAGIGVTAAAGLDLTT--QIWTVPYEVTAELVRSIDRPD
MMAR_3401|M.marinum_M PLTSQVVDYIEAEGIEVMDVISLEVEDNRAVGRLDPMRLPHIAHRLDTHD
**. :..:: *: * . * : : : .. * *
MAB_4413c|M.abscessus_ATCC_199 AEAIFVS-CTNLRTYDVIDPLEQLLGKPVLTANQLTIWACLDRMSLPMMG
MSMEG_2495|M.smegmatis_MC2_155 AEAIFIS-CTNLPTYDVIAKLEEELGKPVLSANQVTIWSALRAAGHKAVG
MMAR_3401|M.marinum_M ADAVVLSACVQMPSLAAIPAAEAMLGLPVISAATATSWKILRSLGVAPVA
*:*:.:* *.:: : .* * ** **::* * * * . :.
MAB_4413c|M.abscessus_ATCC_199 PGRWLRDVFGGGGSSD
MSMEG_2495|M.smegmatis_MC2_155 PGQRLLAS--------
MMAR_3401|M.marinum_M ADAGALFDGGASG---
..