For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SQVSTRLVRLLNMVPYFQANPKVTRAEAAAALGVTGKQLDADLDQLWMCGLPGYSPGDLIDFDFVGDTIE VTFSAGVDHPLRLTSTEATGILVALRALVDVPGMVDPEAARSAIAKIESAVGSQRAVVEGITEDTSAEPG AAATVRTAVRENRALTLEYYSASRDSLATRTVDPIRVVLVGDNSYLEAWCRSAEAVRLFRFDRIVDAQLL DDPAAPPPPAVAAGPDTSLFDADPSLPSATLLIGAAAAWMFDYYPLRDITERPDGSCEATMTYASEDWMA RFILGFGAEVQVLAPESLATRVRQAAEAALQAYARCV*
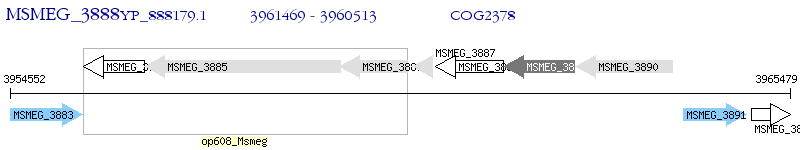
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3888 | - | - | 100% (318) | hypothetical protein MSMEG_3888 |
| M. smegmatis MC2 155 | MSMEG_1359 | - | 1e-10 | 25.39% (323) | DeoR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_3967 | - | 5e-09 | 24.64% (211) | TonB-dependent receptor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2122c | - | 1e-125 | 70.06% (314) | hypothetical protein Mb2122c |
| M. gilvum PYR-GCK | Mflv_3092 | - | 1e-133 | 74.92% (319) | transcriptional regulator-like protein |
| M. tuberculosis H37Rv | Rv2095c | - | 1e-125 | 70.06% (314) | hypothetical protein Rv2095c |
| M. leprae Br4923 | MLBr_01330 | - | 1e-115 | 65.72% (318) | putative DNA-binding protein |
| M. abscessus ATCC 19977 | MAB_2186 | - | 1e-116 | 66.35% (315) | hypothetical protein MAB_2186 |
| M. marinum M | MMAR_3081 | - | 1e-124 | 68.99% (316) | hypothetical protein MMAR_3081 |
| M. avium 104 | MAV_2408 | - | 1e-126 | 70.13% (318) | hypothetical protein MAV_2408 |
| M. thermoresistible (build 8) | TH_0500 | - | 1e-128 | 72.64% (318) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2327 | - | 1e-124 | 68.99% (316) | hypothetical protein MUL_2327 |
| M. vanbaalenii PYR-1 | Mvan_3443 | - | 1e-131 | 73.98% (319) | transcriptional regulator-like protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2122c|M.bovis_AF2122/97 MSALSTRLVRLLNMVPYFQANPRITRAEAAAELGVTAKQLEEDLNQLWMC
Rv2095c|M.tuberculosis_H37Rv MSALSTRLVRLLNMVPYFQANPRITRAEAAAELGVTAKQLEEDLNQLWMC
MMAR_3081|M.marinum_M MSPVSTRLVRLLNMVPYFQANPRITRVEAAAELGVSAKQLEEDLNQLWMC
MUL_2327|M.ulcerans_Agy99 MSPVSTRLVRLLNMVPYFQANPRITRVEAAAELGVSAKQLEEDLNQLWMC
MAV_2408|M.avium_104 MTPISTRLVRLLNMVPYFQANPRVTRAKAAADLGVSAKQLEEDLNQLWMC
MLBr_01330|M.leprae_Br4923 MTQLSTRLVRLLNMVPYFQANPRITRAEAAADLGVSAKQLDQDFNQLWVC
Mflv_3092|M.gilvum_PYR-GCK MTAISARLVRLLNMVPYFQANPRITYTEAAEDLGVSVKQLREDLNQLWMC
Mvan_3443|M.vanbaalenii_PYR-1 MTAVSARLVRLLNMVPYFQANPRITYAEAASDLGVSVKQLREDLNQLWMC
TH_0500|M.thermoresistible__bu MSKASTRLVRLLNMVPYFQANPRITFAEAASDLGVSVKQLREDLTQLWMC
MSMEG_3888|M.smegmatis_MC2_155 MSQVSTRLVRLLNMVPYFQANPKVTRAEAAAALGVTGKQLDADLDQLWMC
MAB_2186|M.abscessus_ATCC_1997 MTELSNRLTRLLNMVPYFQANPGISAAEAAADLGVTTKQLMADLNQLWMC
*: * **.************* :: .:** ***: *** *: ***:*
Mb2122c|M.bovis_AF2122/97 GLPGYSPGDLIDFEFCGDTIEVTFSAGIDRPLKLTSPEATGLLVALRALA
Rv2095c|M.tuberculosis_H37Rv GLPGYSPGDLIDFEFCGDTIEVTFSAGIDRPLKLTSPEATGLLVALRALA
MMAR_3081|M.marinum_M GLPGYFPGDLIDFEFSGDTIEVTFSAGIDRPLKLTSPEATGLLVALRALA
MUL_2327|M.ulcerans_Agy99 GLPGYFPGDLIDFEFSGDTIEVTFSAGIDRPLKLTSPEATGLLVALRALA
MAV_2408|M.avium_104 GLPGYFPGDLIDFEFSGDTIEVTFSAGIDRPLRLTSPEATGLLVALRALA
MLBr_01330|M.leprae_Br4923 GLPGYGPGDLIDFEFSGDTIEVTFSAGIDRPLQLTSPEAIGLLVALRALA
Mflv_3092|M.gilvum_PYR-GCK GLPGYGPGDLIDFEFSGDTIEVTFSAGIDHPLRLTSPEATGVLVALRSLV
Mvan_3443|M.vanbaalenii_PYR-1 GLPGYGPGDLIDFEFSGDTIEVTFSAGIDHPLRLTSPEATGVLVALRALV
TH_0500|M.thermoresistible__bu GLPGYGPGDLIDFDFSGDTVEVTFSAGIDHPLRLTSPEATGILVALRALL
MSMEG_3888|M.smegmatis_MC2_155 GLPGYSPGDLIDFDFVGDTIEVTFSAGVDHPLRLTSTEATGILVALRALV
MAB_2186|M.abscessus_ATCC_1997 GLPGYGPGDLIDLSFSEESIEVTFSAGIDRPLRLTSPEATALLVALRALL
***** ******:.* :::*******:*:**:***.** .:*****:*
Mb2122c|M.bovis_AF2122/97 DIPGVVDPQAARSAIAKIAAAAG--------AVAAVAEQAPTESPAAAAV
Rv2095c|M.tuberculosis_H37Rv DIPGVVDPQAARSAIAKIAAAAG--------AVAAVAEQAPTESPAAAAV
MMAR_3081|M.marinum_M DIPGVVDPQAARSAIAKIAAAAGAAGHSS--TLSAVDEPAPLESPAAAAV
MUL_2327|M.ulcerans_Agy99 DIPGVVDPQAARSAIAKIAAAAGAAEHSS--TLSAVDEPAPLESPAAAAV
MAV_2408|M.avium_104 DIPGVVDPEAARSAIAKIEGAAGAVGHDTTQAATAVDEPAPAESRAAAAV
MLBr_01330|M.leprae_Br4923 NIPGVVDPEAVRSAIAKIEAAAVVMGNEATGSVASVDTRPFSESHAVAAV
Mflv_3092|M.gilvum_PYR-GCK DVPGMVDPEAARSAIAKIESAAGTVG--HGVEGAAVEEPAPLESEAAAAV
Mvan_3443|M.vanbaalenii_PYR-1 DVPGMVDPEAARSAIAKIESAAGKVG--HGVEGAAVEEPAPVESAAAAAV
TH_0500|M.thermoresistible__bu DVPGMVDPAAARSAIAKIEQAAGTVQ--HSAE-TAVDERAPAESAAAAAV
MSMEG_3888|M.smegmatis_MC2_155 DVPGMVDPEAARSAIAKIESAVGSQR--AVVE--GITEDTSAEPGAAATV
MAB_2186|M.abscessus_ATCC_1997 DMPGTVDPEAARSAIAKIEEAVGALP----TDAAPAPLSARTESEAVTAV
::** *** *.******* *. . *. *.::*
Mb2122c|M.bovis_AF2122/97 RAAVRNSRALTIDYYAASHDTLTTRIVDPIRVLLIGGHSYLEAWSREAEG
Rv2095c|M.tuberculosis_H37Rv RAAVRNSRALTIDYYAASHDTLTTRIVDPIRVLLIGGHSYLEAWSREAEG
MMAR_3081|M.marinum_M RSAVLDKRALTIDYYSASHDTLTTRTVDPIRVLLIGGHSYLEAWSRESEG
MUL_2327|M.ulcerans_Agy99 RSAVLDKRALTIDYYSASHDTLTTRTVDPIRVLLIGGHSYLEAWSRESEG
MAV_2408|M.avium_104 RAAVQSRHALAIDYYSASRDTLTSRVVDPIRVLLIGGHSYLEAWSREAEG
MLBr_01330|M.leprae_Br4923 RAAVRDKQALVIDYYSASHDTLTSRIVDPIRVLLVGDHSYLEAWSREAEG
Mflv_3092|M.gilvum_PYR-GCK RTAVRDDRALAIEYYSASHDMLSSRVVDPIRVVLVADHSYLEAWCRTAEA
Mvan_3443|M.vanbaalenii_PYR-1 RKAVRDDRALAIEYYAASHDTMSSRVVDPIRVVLVADHSYLEAWCRSAEA
TH_0500|M.thermoresistible__bu RAATRDKRALALEYYSASHDQLSSRIVDPIRVVLVADHSYLEAWCRSAEG
MSMEG_3888|M.smegmatis_MC2_155 RTAVRENRALTLEYYSASRDSLATRTVDPIRVVLVGDNSYLEAWCRSAEA
MAB_2186|M.abscessus_ATCC_1997 RSAVRNRQALRIEYYSASRDTLSERIVDPIRIAVVGDHSYLEAWCRGSEG
* *. . :** ::**:**:* :: * *****: ::..:******.* :*.
Mb2122c|M.bovis_AF2122/97 VRLFRFDRIVDAAELGEPAVPPESARQAPPDTSLFD---GDLSLPSATLR
Rv2095c|M.tuberculosis_H37Rv VRLFRFDRIVDAAELGEPAVPPESARQAPPDTSLFD---GDLSLPSATLR
MMAR_3081|M.marinum_M VRLFRFDRIVDAVELGEPAMPPEPAVQAPPDTSLFD---GDPALPSATLR
MUL_2327|M.ulcerans_Agy99 VRLFRFDRIVDAVELGEPAMPPEPAVQAPPDTSLFD---GDPALPSATLR
MAV_2408|M.avium_104 VRLFRFDRIVEASELDEPAAPPEPVLHAPPDTSLFD---GDPSLPSATLR
MLBr_01330|M.leprae_Br4923 VRLFRFDRIVVARELDEPAAAPETVRKALPDTSFFD---DDPLLPSATLW
Mflv_3092|M.gilvum_PYR-GCK VRLFRFDRIVDARVLDEPAVPPPPAVQSGPDTSLFDPDTADPSLPTATLL
Mvan_3443|M.vanbaalenii_PYR-1 VRLFRFDRIVDARVLDEPSVPPPPAVQAGPDTSLFDPDTADPSLPVATLL
TH_0500|M.thermoresistible__bu VRLFRFDRIVDAQVLDEPAAPPEPAVQAEPDTSLFEPDPDDPSFQSATLL
MSMEG_3888|M.smegmatis_MC2_155 VRLFRFDRIVDAQLLDDPAAPPPPAVAAGPDTSLFD---ADPSLPSATLL
MAB_2186|M.abscessus_ATCC_1997 VRLFRFDRIEQAEVLDQRAAPPAPAVQTATDTGLFE---ADPSLPAATVI
********* * *.: : .* .. : .**.:*: * : **:
Mb2122c|M.bovis_AF2122/97 VAPSASWMLEYYPIR-ELRQLPDGSCEVVMTYASEDWMTRLLLGFGSDVR
Rv2095c|M.tuberculosis_H37Rv VAPSASWMLEYYPIR-ELRQLPDGSCEVAMTYASEDWMTRLLLGFGSDVR
MMAR_3081|M.marinum_M VAPSASWMFEYYPIR-ELRQLADGSCEVAMTYASEDWMTRLMLGFGSTVQ
MUL_2327|M.ulcerans_Agy99 VAPSASWMFEYYPIR-ELRQLADGSCEVAMTYASEDWMTRLMLGFGSTVQ
MAV_2408|M.avium_104 VAPHASWMFEYYPMR-DVRELPDGSCEAVMTYASEDWMTRLVLGLGSGVQ
MLBr_01330|M.leprae_Br4923 VARSVSWIFEYYPMR-QAHELPDGSFQAVMTYASDAWMTRLVLGFGSAVQ
Mflv_3092|M.gilvum_PYR-GCK IDRSAAWMFDYYPLR-VRTEFPDGSCEATMTYASDQWMTRFVLGFGAAVR
Mvan_3443|M.vanbaalenii_PYR-1 IERSAAWMFDYYPLR-VLREFDDGSCEATMTYASDQWMARFVLGFGSAVR
TH_0500|M.thermoresistible__bu IDRSAGWIFDYYPLR-VLREHDDGSCEAAMTYASDDWMSRLVLGFGSAVR
MSMEG_3888|M.smegmatis_MC2_155 IGAAAAWMFDYYPLR-DITERPDGSCEATMTYASEDWMARFILGFGAEVQ
MAB_2186|M.abscessus_ATCC_1997 VGPSASWVFDYFPMRPTGTQHPDGSLEAAMTYASEDWMARLILGFGSAIR
: ..*:::*:*:* : *** :..*****: **:*::**:*: ::
Mb2122c|M.bovis_AF2122/97 VLAPESLAQRVRDAATAALDAYQAAAPP------
Rv2095c|M.tuberculosis_H37Rv VLAPESLAQRVRDAATAALDAYQAAAPP------
MMAR_3081|M.marinum_M VLEPESLAQRVRAAAATALNAYQAAELA------
MUL_2327|M.ulcerans_Agy99 VLEPESLAQRVRAAAATALNAYQAAELA------
MAV_2408|M.avium_104 VLAPHSLAQRVREAATAALAAYEALS--------
MLBr_01330|M.leprae_Br4923 VQAPEALAYRVRNAAVAALESYQVTAQA------
Mflv_3092|M.gilvum_PYR-GCK VLEPPDLADRVRQTAAAALQAYRVG---IDAPGE
Mvan_3443|M.vanbaalenii_PYR-1 VLEPVDLADRVRRSAAAALEAYRVGGVGIDPADG
TH_0500|M.thermoresistible__bu VLAPESLARRVAESARAALAVYDLS---AEHAGR
MSMEG_3888|M.smegmatis_MC2_155 VLAPESLATRVRQAAEAALQAYARCV--------
MAB_2186|M.abscessus_ATCC_1997 VLAPESLARRVRADATAALAAYQAS---------
* * ** ** * :** *