For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MARTVDTTGRVVQLLGLLQSRRVWTGEELAERLGVTGRSVRRDIERLRELGYPVHASKGQGGGYQLGAGM ALPPLLLDPDEAVAMAVCLRLAAGGSVAGVGESALRALSKLDQVMPARLRSQVAAIHDATVTLGPNATDT AVAPDVLMTLARACRDREHVSTGYTDLRGNQTQRRLEPYQLVTTGRRWYLMAYDRDREDWRSLRLDRMSD VRATGTTFTARPAPDAAAYVGRAISASAYPYVARVRYFAPEKVVAQRFPPGTATFEPDGPDACIVTSGAE YPEQLAMYFATVGHDFEVLEPAEVIDAVGAMADRLRRAVR
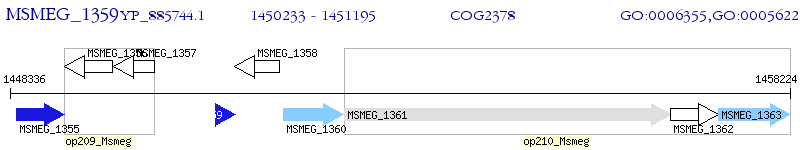
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1359 | - | - | 100% (320) | DeoR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_3967 | - | 2e-15 | 29.58% (213) | TonB-dependent receptor |
| M. smegmatis MC2 155 | MSMEG_3888 | - | 1e-10 | 25.39% (323) | hypothetical protein MSMEG_3888 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2123c | - | 7e-13 | 26.17% (321) | hypothetical protein Mb2123c |
| M. gilvum PYR-GCK | Mflv_5106 | - | 1e-134 | 76.36% (313) | helix-turn-helix, type 11 domain-containing protein |
| M. tuberculosis H37Rv | Rv2096c | - | 7e-13 | 26.17% (321) | hypothetical protein Rv2096c |
| M. leprae Br4923 | MLBr_01329 | - | 5e-14 | 28.13% (327) | hypothetical protein MLBr_01329 |
| M. abscessus ATCC 19977 | MAB_2185 | - | 1e-13 | 28.42% (285) | hypothetical protein MAB_2185 |
| M. marinum M | MMAR_3082 | - | 6e-11 | 27.62% (315) | hypothetical protein MMAR_3082 |
| M. avium 104 | MAV_2407 | - | 4e-13 | 28.40% (324) | hypothetical protein MAV_2407 |
| M. thermoresistible (build 8) | TH_1858 | - | 1e-136 | 75.40% (313) | DeoR-family protein transcriptional regulator |
| M. ulcerans Agy99 | MUL_2328 | - | 6e-12 | 27.62% (315) | hypothetical protein MUL_2328 |
| M. vanbaalenii PYR-1 | Mvan_1247 | - | 1e-135 | 76.51% (315) | helix-turn-helix, type 11 domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3082|M.marinum_M ----MATSKVERMVNLVIALLSTRGYITAEKVRSSVAGYSDSPTAEAFSR
MUL_2328|M.ulcerans_Agy99 ----MATSKVERMVNLVIALLSTRGYITAEKVRSSVAGYSDSPTAEAFSR
Mb2123c|M.bovis_AF2122/97 ----MATSKVERLVNLVIALLSTRGYITAEKIRSSVAGYSDSPSVEAFSR
Rv2096c|M.tuberculosis_H37Rv ----MATSKVERLVNLVIALLSTRGYITAEKIRSSVAGYSDSPSVEAFSR
MAV_2407|M.avium_104 --MPMATSKVERLVNLVIALLSTRGYITAEKIRSSVAGYSESASAEAFSR
MLBr_01329|M.leprae_Br4923 ----MATSKVERLVNLVIALLSTRGYMTAEKIRSSVAGYSDSPTVEAFSR
MAB_2185|M.abscessus_ATCC_1997 ----MAISKVERLMNLVICLLSTRTYVTAERIRTSVAGYSDSPSDEAFSR
Mflv_5106|M.gilvum_PYR-GCK ------MSETTGRVLQLLGLLQSRRVWSGDELAARLR---------VTTR
Mvan_1247|M.vanbaalenii_PYR-1 ------MSETTGRVLQLLGLLQSRRVWSGEELAGRLG---------VTTR
TH_1858|M.thermoresistible__bu MWQAADVSETTSRVLRLLGLLQSRRVWSGEELAERLG---------VTTR
MSMEG_1359|M.smegmatis_MC2_155 ---MARTVDTTGRVVQLLGLLQSRRVWTGEELAERLG---------VTGR
.. : :: **.:* :.:.: : . *
MMAR_3082|M.marinum_M MFERDKNELRDLGIPLEVGRVSSTDPTEGYRINRDAYALPPVELTPDETA
MUL_2328|M.ulcerans_Agy99 MFERDKNELRDLGIPLEVGRVSSTDPTEGYRINRDAYALPPVELTPDETA
Mb2123c|M.bovis_AF2122/97 MFERDKNELRDLGIPLEVGRVSALEPTEGYRINRDAYALSPVELTPDEAA
Rv2096c|M.tuberculosis_H37Rv MFERDKNELRDLGIPLEVGRVSALEPTEGYRINRDAYALSPVELTPDEAA
MAV_2407|M.avium_104 MFERDKNELRDLGIPLEVGRVSPMDPTEGYRINRDAYALAPVDLTPDEAA
MLBr_01329|M.leprae_Br4923 MFERDKNELRDLGIPLEVGKVSALDPSEGYRINRDAYALPPVELTPDEAA
MAB_2185|M.abscessus_ATCC_1997 MFERDKNELRDLGIPLETGKVSSFDATEGYRIRRDAYELPDITLTADESA
Mflv_5106|M.gilvum_PYR-GCK SVRRDIDRLRELGYPVHAS----TGHGGGYQLG-AGAALPPLLLDPDEAV
Mvan_1247|M.vanbaalenii_PYR-1 SVRRDVDRLRELGYPVHAS----TGHGGGYQLG-AGAALPPLLLDPEEAV
TH_1858|M.thermoresistible__bu SVRRDVERLRDLGYPVHAS----RGHGGGYQLG-AGAALPPLLLDPEEAV
MSMEG_1359|M.smegmatis_MC2_155 SVRRDIERLRELGYPVHAS----KGQGGGYQLG-AGMALPPLLLDPDEAV
..** :.**:** *:... **:: . *. : * .:*:.
MMAR_3082|M.marinum_M AVAVATQLWESPELITATQGALLKLRAAGVDVDP---DAPVAIASPAGVP
MUL_2328|M.ulcerans_Agy99 AVAVATQLWESPELITATQGALLKLRAAGVDVDP---DAPVAIASPAGVP
Mb2123c|M.bovis_AF2122/97 AVAVATQLWESPELITATQGALLKLRAAGVDVDPLDTGAPVAIASAAAVS
Rv2096c|M.tuberculosis_H37Rv AVAVATQLWESPELITATQGALLKLRAAGVDVDPLDTGAPVAIASAAAVS
MAV_2407|M.avium_104 AVAVATQLWESPELITATQGALLKLRAAGVDVDPLDDGAPVAIASPAGVP
MLBr_01329|M.leprae_Br4923 AVAVATQLWESQELITATQGALLKLRAAGVDIDP--LDTPVVIASSSGVS
MAB_2185|M.abscessus_ATCC_1997 AVAVATQLWQSPELVTAAQGALMKLRAAGVDVDP--AAENVSVAAPAAMP
Mflv_5106|M.gilvum_PYR-GCK AVAVCLRVAAGGSVAGVGESALRALSKLDQVMPSRLRSQVAAVHDTTVTL
Mvan_1247|M.vanbaalenii_PYR-1 AMAVCLRVAAGGSVAGVGESALRALSKLDQVMPARLRSQVAAVHDATVTL
TH_1858|M.thermoresistible__bu AVAVCLRLAAGGSIAGVGESALRALGKLDQVMPARLRSQVSAVHEATTTL
MSMEG_1359|M.smegmatis_MC2_155 AMAVCLRLAAGGSVAGVGESALRALSKLDQVMPARLRSQVAAIHDATVTL
*:**. :: . .: . :.** * . : . : .:
MMAR_3082|M.marinum_M GLRGSE-----DVLGILLAAIDSGQAVRFPHRASRHEPYVTRTVEPWGVV
MUL_2328|M.ulcerans_Agy99 GLRGSE-----DVLGILLAAIDSGQAVRFPHRASRHEPYVTRTVEPWGVV
Mb2123c|M.bovis_AF2122/97 GLRGSE-----DVLGILLSAIDSGQVVQFSHRSSRAEPYTVRTVEPWGVV
Rv2096c|M.tuberculosis_H37Rv GLRGSE-----DVLGILLSAIDSGQVVQFSHRSSRAEPYTVRTVEPWGVV
MAV_2407|M.avium_104 GLRGSE-----EVLGILLSAIDSRQAVQFPHRPSRAEPYATRTVEPWGVV
MLBr_01329|M.leprae_Br4923 SLRGSE-----DFLSILLSAIGSRQAVQFPYRPSRAEPYTMRNVEPWGVI
MAB_2185|M.abscessus_ATCC_1997 GGRGSE-----SVLRILLSAIDSGQAVQFGHRPSRTEPYVTRTVEPWGVI
Mflv_5106|M.gilvum_PYR-GCK A-GATDTPVEPEVLMTLARASRDHEHVTVGYTDVRGN-VTSRRVEPYQLV
Mvan_1247|M.vanbaalenii_PYR-1 G-AATDTPVEPDVLMTLARASRDHEHVTADYVDIRGN-VTQRRLEPYHLV
TH_1858|M.thermoresistible__bu GPNQPDQPVAPDVLMTLARACRDHEHVNTGYTDRAGN-RSRRRLEPYQLV
MSMEG_1359|M.smegmatis_MC2_155 GPNATDTAVAPDVLMTLARACRDREHVSTGYTDLRGN-QTQRRLEPYQLV
. .: ..* * * . : * : : * :**: ::
MMAR_3082|M.marinum_M TEKGRWYLVGHDRDRGATRTFRLSRIGADVAPVGAPGEAEVPEDVDLRQI
MUL_2328|M.ulcerans_Agy99 TEKGRWYLVGHDRDRGATRTFRLSRIGADVAPVGAPGESKIPEDVDLRQI
Mb2123c|M.bovis_AF2122/97 TEKGRWYLVGHDRDRDATRVFRLSRIGAQVTPIGPAGATTVPAGVDLRSI
Rv2096c|M.tuberculosis_H37Rv TEKGRWYLVGHDRDRDATRVFRLSRIGAQVTPIGPAGATTVPAGVDLRSI
MAV_2407|M.avium_104 TQKGRWYLVGHDRDRDATRTFRLSRIGPGVTPIGPPGAVTVPDGVDLRRI
MLBr_01329|M.leprae_Br4923 TENSCWYLVGHDCDRNATRTFRLSRIGSEVAPIGPAGAVTVPDGVDLRRI
MAB_2185|M.abscessus_ATCC_1997 TDRGRWYLVGHDRDRDATRTFRLSRIGEDVTKIEKPGTIRRPEGADLRAI
Mflv_5106|M.gilvum_PYR-GCK TTGRRWYLMCFDRDRDDWRSLRLDRM-SDVRAVG--TTFIPREAPDAASY
Mvan_1247|M.vanbaalenii_PYR-1 TTGRRWYLMCFDRDRVDWRSLRLDRM-SQVRAVG--STFTPRDAPDAASY
TH_1858|M.thermoresistible__bu TTGRRWYLLAYDRDRGDWRSLRLDRM-TDVRALG--STFTPRPAPDAAEF
MSMEG_1359|M.smegmatis_MC2_155 TTGRRWYLMAYDRDREDWRSLRLDRM-SDVRATG--TTFTARPAPDAAAY
* ***: .* ** * :**.*: * *
MMAR_3082|M.marinum_M VARTVAES---PTGMQATVWVADGRATALRRAGRSMGPRRLAGRDGEVIE
MUL_2328|M.ulcerans_Agy99 VARTVAES---PTGMQATVWVADGRATALRRAGPSMGPRRLAGRDGEVIK
Mb2123c|M.bovis_AF2122/97 VAQKVTEV---PTGEQATVWVAEGRATALRRAGRSAGPRQLGGRDGEVIE
Rv2096c|M.tuberculosis_H37Rv VAQKVTEV---PTGEQATVWVAEGRATALRRAGRSAGPRQLGGRDGEVIE
MAV_2407|M.avium_104 VAQTVTDVSATPAGGQARVWVADGRATALRRAGRPVGTRQLGGRDGEVIE
MLBr_01329|M.leprae_Br4923 VSDAVAEV---STGATARVWVVDGRATALRHAGRPAGVRRLGGRDGQVIE
MAB_2185|M.abscessus_ATCC_1997 VAAAVGDG---PVRITARVWIAEGRAQELRRMGVVVDSRRLGERDGDIVE
Mflv_5106|M.gilvum_PYR-GCK VRRAISSS---PYPYVARVRYRAPADVVARVFPAASVDVEPDGPDACIVT
Mvan_1247|M.vanbaalenii_PYR-1 VRRSISSS---PYPYIARVRYFAPHDVVAQAFSAASVHIEPEGPDACILT
TH_1858|M.thermoresistible__bu VRHSISAS---PYRHVARVRFRAPERVVSQHFPAAAVMIEPDGPDTCIVT
MSMEG_1359|M.smegmatis_MC2_155 VGRAISAS---AYPYVARVRYFAPEKVVAQRFPPGTATFEPDGPDACIVT
* : . * * : . * ::
MMAR_3082|M.marinum_M LEIGSSDRLARDITGHGADAIVLEPPALREDVLARLRALAGVEEALA---
MUL_2328|M.ulcerans_Agy99 LEIGSSDRLARDITGHGADAIVLEPPALREDVLARLRALAGVEEALA---
Mb2123c|M.bovis_AF2122/97 LEIRSSDRLAREITGYGADAIVLQPGSLRDDVLARLRAQAGALA------
Rv2096c|M.tuberculosis_H37Rv LEIRSSDRLAREITGYGADAIVLQPGSLRDDVLARLRAQAGALA------
MAV_2407|M.avium_104 LDIRATDQLARDIAGYGADAVVLEPQSLRDDVLARLRAHAGADAVGAGRP
MLBr_01329|M.leprae_Br4923 LDIGSIDRLARDIAGHGADAVVLEPDALRDDVLIRLRAHAGTGPS-----
MAB_2185|M.abscessus_ATCC_1997 IDVASPDRLVRSIASHGADAVALEPAQVRDEVVARLQEQAGVTA------
Mflv_5106|M.gilvum_PYR-GCK VGADDPQRMVPWLAMPGCEFEVLEPDEVVDAVRVVAERIARAGAGGD---
Mvan_1247|M.vanbaalenii_PYR-1 AGADDPERMVPWLAMPGCDFEVLEPPEVIEAAQRVAGRIARACRT-----
TH_1858|M.thermoresistible__bu AGADDPQRIVLHLAAPGLPFEVLEPAEVAEAAAAVADLLTRASGRT----
MSMEG_1359|M.smegmatis_MC2_155 SGAEYPEQLAMYFATVGHDFEVLEPAEVIDAVGAMADRLRRAVR------
:::. :: * .*:* : : . .
MMAR_3082|M.marinum_M --------
MUL_2328|M.ulcerans_Agy99 --------
Mb2123c|M.bovis_AF2122/97 --------
Rv2096c|M.tuberculosis_H37Rv --------
MAV_2407|M.avium_104 GGRGGVSA
MLBr_01329|M.leprae_Br4923 --------
MAB_2185|M.abscessus_ATCC_1997 --------
Mflv_5106|M.gilvum_PYR-GCK --------
Mvan_1247|M.vanbaalenii_PYR-1 --------
TH_1858|M.thermoresistible__bu --------
MSMEG_1359|M.smegmatis_MC2_155 --------