For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MADHVAMTQLEVADYDLGLRGKLVRSSKTHGASGLAFCTILYGLSLIDDVTDTPFSNAANGYPDAQLVPD ETTRVALPWRHGTDAVIAELVDEQGRPLEVSPRAVLAGLAARYGELGLAPVLGYEYELWIFRDGEGTSPP ILGGRPAHGRTENAYSLTRCAEINDIATEFVNRMDQIGIEVEMFHAELGPGFFEFTLAPETALRATDNAA RARQYLRDLCAERGLHASFMAKPFADKSGAGGHVHSSLTRDGNNVFSDGAGALSAEGSRYLAGLLAGMAD TSLMLNPHVNSYKRIDPEMFTPALANWGYDDRGTSARVILPDAKSARVEHRRPGADASPYLVAAALLAAG LRGLQEELPLPERDAEPTELPGDLAGALAHFENSTWLPDLLGKAFCTSYAATRRAELQRYEQWLRTTITD WELHRHLEHQ
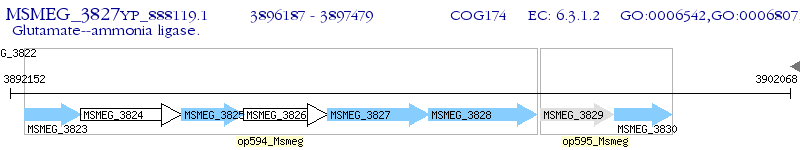
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3827 | - | - | 100% (430) | glutamine synthetase |
| M. smegmatis MC2 155 | MSMEG_2595 | - | 6e-38 | 29.44% (411) | gamma-glutamylisopropylamide synthetase |
| M. smegmatis MC2 155 | MSMEG_1116 | - | 1e-37 | 31.27% (403) | gamma-glutamylisopropylamide synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2885c | glnA4 | 4e-39 | 29.90% (408) | glutamine synthetase |
| M. gilvum PYR-GCK | Mflv_2909 | - | 8e-39 | 31.54% (390) | glutamine synthetase, type I |
| M. tuberculosis H37Rv | Rv2860c | glnA4 | 4e-39 | 29.90% (408) | glutamine synthetase |
| M. leprae Br4923 | MLBr_01631 | glnA2 | 9e-35 | 30.49% (387) | glutamine synthase class II |
| M. abscessus ATCC 19977 | MAB_1920 | - | 9e-37 | 30.75% (387) | glutamine synthetase |
| M. marinum M | MMAR_3294 | glnA2 | 5e-35 | 30.00% (390) | glutamine synthetase GlnA2 |
| M. avium 104 | MAV_3718 | glnA | 1e-42 | 31.95% (410) | glutamine synthetase catalytic domain |
| M. thermoresistible (build 8) | TH_3396 | - | 3e-38 | 31.27% (387) | PUTATIVE glutamine synthetase, type I |
| M. ulcerans Agy99 | MUL_1335 | glnA2 | 4e-35 | 30.00% (390) | glutamine synthetase GlnA2 |
| M. vanbaalenii PYR-1 | Mvan_0611 | - | 1e-174 | 70.42% (426) | glutamate--ammonia ligase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2909|M.gilvum_PYR-GCK -----------------MDRQKEFVLRTLEERDIRFVRLWFTDVLGYLKS
TH_3396|M.thermoresistible__bu -----------------MDRQKEFVLRSLEERDIRFVRLWFTDVLGFLKS
MMAR_3294|M.marinum_M -----------------MDRQKEFVLRTLEERDIRFVRLWFTDVLGFLKS
MUL_1335|M.ulcerans_Agy99 -----------------MDRQKEFVLRTLEERDIRFVRLWFTDVLGFLKS
MLBr_01631|M.leprae_Br4923 -----------------MDRQKEFVLRTLEERDIRFVRLWFTDVLGYLKS
MAB_1920|M.abscessus_ATCC_1997 -----------------MDRQKEFVLRTLEERDIRFVRLWFTDVLGYLKS
Mb2885c|M.bovis_AF2122/97 -------MTGPGSPPLAWTELERLVAAGD----VDTVIVAFTDMQGRLAG
Rv2860c|M.tuberculosis_H37Rv -------MTGPGSPPLAWTELERLVAAGD----VDTVIVAFTDMQGRLAG
MAV_3718|M.avium_104 MASEDSALTGSDTAMLSLAALDRLVAAGAPETRVDTVIVAFPDMQGRLVG
MSMEG_3827|M.smegmatis_MC2_155 -----------------------------MADHVAMTQLEVADYDLGLRG
Mvan_0611|M.vanbaalenii_PYR-1 -------------------------------MTVPMTQLEVPDYDLGIRG
: . : ..* : .
Mflv_2909|M.gilvum_PYR-GCK VAIAPAELEGAFEEG----------IGFDGSSIEGFARVSEA----DMVA
TH_3396|M.thermoresistible__bu VAIAPAELEGAFEEG----------IGFDGSAIEGFARVSEA----DMVA
MMAR_3294|M.marinum_M VAIAPAELEGAFEEG----------IGFDGSSIEGFARVSES----DTVA
MUL_1335|M.ulcerans_Agy99 VAIAPAELEGAFEEG----------IGFDGSSIEGFARVSES----DTVA
MLBr_01631|M.leprae_Br4923 VAIAPAELEGAFEEG----------IGFDGSSIEGFARVSES----DTVA
MAB_1920|M.abscessus_ATCC_1997 VSIAPAELEGAFEEG----------IGFDGSSIEGFARVSES----DTVA
Mb2885c|M.bovis_AF2122/97 KRISGRHFVDDIATRGVECCSYLLAVDVDLNTVPGYAMASWDTGYGDMVM
Rv2860c|M.tuberculosis_H37Rv KRISGRHFVDDIATRGVECCSYLLAVDVDLNTVPGYAMASWDTGYGDMVM
MAV_3718|M.avium_104 KRMDARLFVDEAAATGVECCGYLLAVDVDMNTVGGYAISGWDTGFGDLVM
MSMEG_3827|M.smegmatis_MC2_155 KLVRSSKTHGASGLAFCTILYGLSLIDDVTDTPFSNAANGYP----DAQL
Mvan_0611|M.vanbaalenii_PYR-1 KLVRTEKTVDPTRLAFCTILYGLSVVDEVSDTPVSNAGNGYP----DARL
: . :. .: . * . *
Mflv_2909|M.gilvum_PYR-GCK RPDPSTFQVLPWADSSGKHHSARMFCDITMPDGSPSWADSRHVLRRQLSK
TH_3396|M.thermoresistible__bu RPDPSTFQVLPWTGDSGKHYSARMFCDITMPDGSPSWADPRHVLRRQLAK
MMAR_3294|M.marinum_M HPDPSTFQVLPWVTDAGHHHSARIFCDITMPDGSPSWADPRHVLRRQLTK
MUL_1335|M.ulcerans_Agy99 HPDPSTFQVLPWVTDAGHHHSARIFCDITMPDGSPSWADPRHVLRRQLIK
MLBr_01631|M.leprae_Br4923 HPDPSTFQILPWATTAGHHHSARMFCDITMPDGSSSWADPRHVLRRQLTK
MAB_1920|M.abscessus_ATCC_1997 HPDPSTFQVLPWTTSAGKHHSARMFCDIKLPDGSPSWSDPRHVLRRQLAK
Mb2885c|M.bovis_AF2122/97 TPDLSTLRLIPWLPG-----TALVIADLVWADGSEVAVSPRSILRRQLDR
Rv2860c|M.tuberculosis_H37Rv TPDLSTLRLIPWLPG-----TALVIADLVWADGSEVAVSPRSILRRQLDR
MAV_3718|M.avium_104 RPDLSTLRRIPWLPG-----TALVIADVVGADGSPVAVSPRAVLRRQLDR
MSMEG_3827|M.smegmatis_MC2_155 VPDETTRVALPWRHG-----TDAVIAELVDEQGRPLEVSPRAVLAGLAAR
Mvan_0611|M.vanbaalenii_PYR-1 VPDQSTRVALPWRPG-----TDAVIADMVGADGAPLAMSPRNSLRSLVDR
** :* :** : ::.:: :* ..* * :
Mflv_2909|M.gilvum_PYR-GCK ASDLGFSCYVHPEIEFFLLKPGDNDG--SPPTPADNGGYFDQAVHDSAPN
TH_3396|M.thermoresistible__bu AGDQGFTCYVHPEIEFFLLKPGPHDG--SPPVPADTSGYFDQSVHDTAPN
MMAR_3294|M.marinum_M ANELGFSCYVHPEIEFFLLNPGPEDG--TEPVPIDNAGYFDQAVHVSASK
MUL_1335|M.ulcerans_Agy99 ANELGFSCYVHPEIEFFLLNPGPEDG--TEPVPIDNAGYFDQAVHVSASK
MLBr_01631|M.leprae_Br4923 ANDLGFSCYVHPEIEFFLLKAGPEDGSMPIPVPVDNAGYFDQAVHDSALN
MAB_1920|M.abscessus_ATCC_1997 ASDLGFSCYVHPEIEFFLLKNLPDDG--TAPIPADDAGYFDQAVHEAAPN
Mb2885c|M.bovis_AF2122/97 LKARGLVADVATELEFIVFDQPYRQAWASGYRGLTPASDYNIDYAILASS
Rv2860c|M.tuberculosis_H37Rv LKARGLVADVATELEFIVFDQPYRQAWASGYRGLTPASDYNIDYAILASS
MAV_3718|M.avium_104 LAGRGLFADAATELEFMVFDEPYRQAWASGYRGLTPASDYNIDYAISASS
MSMEG_3827|M.smegmatis_MC2_155 YGELGLAPVLGYEYELWIFRDG-EGTSPPILGGRPAHGRTENAYSLTRCA
Mvan_0611|M.vanbaalenii_PYR-1 YAELDLEPVLGFEYEFWLFQDR-EDP-------RRPLGSTENAYSLSRIA
.: * *: :: . :
Mflv_2909|M.gilvum_PYR-GCK ----FRRHAIDALEQMGISVEFSHHEGAPGQQEIDLRYADALSMADNVMT
TH_3396|M.thermoresistible__bu ----FRRHAIEALEQMGISVEFSHHEGAPGQQEIDLRYADALSMADNVMT
MMAR_3294|M.marinum_M ----FRRHAIDALEFMGISVEFSHHEGAPGQQEIDLRFADALSMADNVMT
MUL_1335|M.ulcerans_Agy99 ----FRRHVIDALEFMGISVEFSHHEGAPGQQEIDLRFADALSMADNVMT
MLBr_01631|M.leprae_Br4923 ----FRRHAIEALEFMGISVEFSHHEGAPGQQEIDLRFADALSMADNVMT
MAB_1920|M.abscessus_ATCC_1997 ----FRRHAIDALESMGISVEFSHHEGAPGQQEIDLRYADALSMADNVMT
Mb2885c|M.bovis_AF2122/97 RMEPLLRDIRLGMAGAGLRFEAVKGECNMGQQEIGFRYDEALVTCDNHAI
Rv2860c|M.tuberculosis_H37Rv RMEPLLRDIRLGMAGAGLRFEAVKGECNMGQQEIGFRYDEALVTCDNHAI
MAV_3718|M.avium_104 RMEPLLRDIRRGMAGAGLRFESVKGECNRGQQEIGFRYDEALRTCDNHVI
MSMEG_3827|M.smegmatis_MC2_155 EINDIATEFVNRMDQIGIEVEMFHAELGPGFFEFTLAPETALRATDNAAR
Mvan_0611|M.vanbaalenii_PYR-1 ESQSLATEFIDRMHAVGIEIEMFHTELGPGFFEFTMAPAPACQAADAAVR
: . : *: .* : * * *: : * *
Mflv_2909|M.gilvum_PYR-GCK FRYLVKEVALGDGVRASFMPKPFAEHPGSAMHTHMSLFEGD-SNAFHSPD
TH_3396|M.thermoresistible__bu FRYVVKQVAISDGVRASFMPKPFAEHPGSAMHTHMSLFEGD-TNAFHSPD
MMAR_3294|M.marinum_M FRYVIKEVALEEGVRATFMPKPFGQHAGSAMHTHMSLFEGD-VNAFHSPD
MUL_1335|M.ulcerans_Agy99 FRYVIKEVALEEGVRATFMPKPFGQHAGSAMHTHMSLFEGD-VNAFHSPD
MLBr_01631|M.leprae_Br4923 FRYVIKEVAIENGARASFMPKPFAQHPGSAMHTHMSLFEGD-VNAFHSDD
MAB_1920|M.abscessus_ATCC_1997 FRNVVKEVAIIDGVHATFMPKPFSDHPGSAMHTHMSLFEGD-TNAFHNPD
Mb2885c|M.bovis_AF2122/97 YKNGAKEIADQHGKSLTFMAK-YDEREGNSCHIHVSLRGTDGSAVFADSN
Rv2860c|M.tuberculosis_H37Rv YKNGAKEIADQHGKSLTFMAK-YDEREGNSCHIHVSLRGTDGSAVFADSN
MAV_3718|M.avium_104 YKNGAKEIADQHGKSLTFMAK-YDEREGNSCHVHLSLRDAQGGAAFADPS
MSMEG_3827|M.smegmatis_MC2_155 ARQYLRDLCAERGLHASFMAKPFADKSGAGGHVHSSLTRDG-NNVFSDGA
Mvan_0611|M.vanbaalenii_PYR-1 ARQYLRDLCAERGLHASFMAKPFGDKSGAGGHVHSSLNRNG-KNIFADDH
: :::. * :**.* : :: * . * * ** * .
Mflv_2909|M.gilvum_PYR-GCK DPLQLSAVGKSFIAGILEHANEISAVTNQWVNSYKRLVHGGEAPTAASWG
TH_3396|M.thermoresistible__bu DPLQLSDVAKSFIAGILEHANEISAVTNQWVNSYKRLVHGGEAPTAASWG
MMAR_3294|M.marinum_M DPLQLSEVGKSFIAGILEHASEISAVTNQWVNSYKRLILGGEAPTAASWG
MUL_1335|M.ulcerans_Agy99 DPLQLSEVGKSFIAGILEHASEISAVTNQWVNSYKRLILGGEAPTAASWG
MLBr_01631|M.leprae_Br4923 DPLQLSDVGKSFIAGILEHASEISAVTNQWVNSYKRLVVGGEAPTTASWG
MAB_1920|M.abscessus_ATCC_1997 DPLQLSDVGKSFIAGILEHANEISAVTNQWVNSYKRLVQGGEAPTAASWG
Mb2885c|M.bovis_AF2122/97 GPHGMSSMFRSFVAGQLATLREFTLCYAPTINSYKRFADSSFAPTALAWG
Rv2860c|M.tuberculosis_H37Rv GPHGMSSMFRSFVAGQLATLREFTLCYAPTINSYKRFADSSFAPTALAWG
MAV_3718|M.avium_104 RPHGMSTMFCSFLAGLLATMADFTLFYAPNINSYKRFADESFAPTALAWG
MSMEG_3827|M.smegmatis_MC2_155 G--ALSAEGSRYLAGLLAGMADTSLMLNPHVNSYKRIDPEMFTPALANWG
Mvan_0611|M.vanbaalenii_PYR-1 G--WLSAEAGHYLAGLLAGMADTTAMLNPYVNSFKRIDPEMFTPVHATYG
:* ::** * : : :**:**: :*. :*
Mflv_2909|M.gilvum_PYR-GCK AANRSALVRVPMYTPHKVSSRRVEVRSPDSACNPYLTFAVLLAAGLRGIE
TH_3396|M.thermoresistible__bu AANRSALVRVPMYTPHKASSRRVEVRSPDSACNPYLTFAVLLAAGLRGVE
MMAR_3294|M.marinum_M AANRSALVRVPMYTPHKTSSRRIEVRSPDSACNPYLAFAVLLAAGLRGVE
MUL_1335|M.ulcerans_Agy99 AANRSALVRVPMYTPHKTSSRRIEVRSPDSACNPYLAFAVLLAAGLRGVE
MLBr_01631|M.leprae_Br4923 AANRSALVRVPMYTPHKTSSRRVEVRSPDSACNPYLAFAVLLAAGLRGVE
MAB_1920|M.abscessus_ATCC_1997 AANRSALVRVPMYTPNKSSSRRIEVRSPDSACNPYLTYAVLLAAGLRGVE
Mb2885c|M.bovis_AF2122/97 LDNRTCALRVVGHGQN----IRVECRVPGGDVNQYLAVAALIAGGLYGIE
Rv2860c|M.tuberculosis_H37Rv LDNRTCALRVVGHGQN----IRVECRVPGGDVNQYLAVAALIAGGLYGIE
MAV_3718|M.avium_104 LDNRTCALRVVGHGAH----TRVECRVPGGDVNPYLAVAAIVAGGLYGIE
MSMEG_3827|M.smegmatis_MC2_155 YDDRGTSARVILPDAKS---ARVEHRRPGADASPYLVAAALLAAGLRGLQ
Mvan_0611|M.vanbaalenii_PYR-1 HDDRSTACRLILNDAKS---ARVEHRRPGADASPYLVATALLAAGFLGLQ
:* *: : *:* * *.. . **. :.::*.*: *::
Mflv_2909|M.gilvum_PYR-GCK KNYVLGPEAEDNVWSLTPEERRAMGYKELPGSLGAALADMENSELVAEAL
TH_3396|M.thermoresistible__bu KGYVLGPEAEDNVWTLTPEERRAMGFKELPSSLGQALAEMERSELVAEAL
MMAR_3294|M.marinum_M KGYVLGPQAEDNVWDLTPEERRTMGYRELPTSLDSALRAMESSELVAEAL
MUL_1335|M.ulcerans_Agy99 KGYVLGPQAEDNVWDLTPEERRTMGYRELPTSLDSALRAMESSELVAEAL
MLBr_01631|M.leprae_Br4923 KGYVLGPQAEDNVWDLTPEERLAMGYRELPTSLDSALGAMEVSEFVAEAL
MAB_1920|M.abscessus_ATCC_1997 QGYTLGPEAEDDVWGLTRAERQAMGYKELPSTLENALIAMEGSELVAEAL
Mb2885c|M.bovis_AF2122/97 RGLQLPEPCVGNAYQGADVER-------LPVTLADAAVLFEDSALVREAF
Rv2860c|M.tuberculosis_H37Rv RGLQLPEPCVGNAYQGADVER-------LPVTLADAAVLFEDSALVREAF
MAV_3718|M.avium_104 QGLALPEPCAGNAYRARGVGR-------LPATLAEAAALFEHSALARQVF
MSMEG_3827|M.smegmatis_MC2_155 EELPLPERDAEPTE--------------LPGDLAGALAHFENSTWLPDLL
Mvan_0611|M.vanbaalenii_PYR-1 ENLTLPGSDAVPAP--------------LPGDLTAAIDAFENSPWLPDLV
. * . ** * * :* * : .
Mflv_2909|M.gilvum_PYR-GCK GEHVFDFFLRNKRREWENYR----SHVTPFELKNYLSL-
TH_3396|M.thermoresistible__bu GEHVFDFFLRNKRAEWENYR----SHVTPYELKNYLSL-
MMAR_3294|M.marinum_M GEHVFDFFLRNKRREWATYR----SHVTPYELSTYLSL-
MUL_1335|M.ulcerans_Agy99 GEHVFDFFLRNKRREWATYR----SHVTPYELSTYLSL-
MLBr_01631|M.leprae_Br4923 GEHVFDFFLRNKRAEWANYR----SHVSLYELKTYLSL-
MAB_1920|M.abscessus_ATCC_1997 GEHVFDFFLRNKRAEWARYR----SNVTPFELRAYLGL-
Mb2885c|M.bovis_AF2122/97 GEDVVAHYLNNARVELAAFN----AAVTDWERIRGFERL
Rv2860c|M.tuberculosis_H37Rv GEDVVAHYLNNARVELAAFN----AAVTDWERIRGFERL
MAV_3718|M.avium_104 GDDVVAHYLNNARVELAAFH----AAVTDWERMRGFERL
MSMEG_3827|M.smegmatis_MC2_155 GKAFCTSYAATRRAELQRYEQWLRTTITDWELHRHLEHQ
Mvan_0611|M.vanbaalenii_PYR-1 GKAFCASFAATRRAEAQRYAQWLKTTITSWELTRHLEHQ
*. . : . * * : : :: :* :