For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSRNPGMLTQDELVQLVTDGEIDTVIVAFCDMQGRLTGKRVSARLFVEEVAANGCECCNYLLAVDVDMNT VDGYAVSSWETGYGDMVMTPDFSTLRRIPWLPGTAMVMADLSWLDGNPVAVAPRSILNHQIDRLAERGLV PFVGTELEFMVFDDSFRQAWSSGYHNLTPATDYNVDYAIHASTRMEPLLRDIRRGMESAGMYCEGVKGEC NLGQQEIAFRYDHARVTCDNHTIYRNGAKEIADQHGKSLTFMAKYDEREGNSCHIHISLRGAGENGDAVF ADESDPHGMSPLFRSFIAGILATLRELTLFYAPNINSYKRFVDGSFAPTAVAWGFDNRTCALRVVGHGHG MRVECRAPGGDVNQYLAVSALIAGGLYGIERDLELPEPMAGNAYTSGAERLPTTLAEAADLFAKSEVARA AFGDDVVEHYLNYARVEMAAFNAAVTDWERVRGFERL
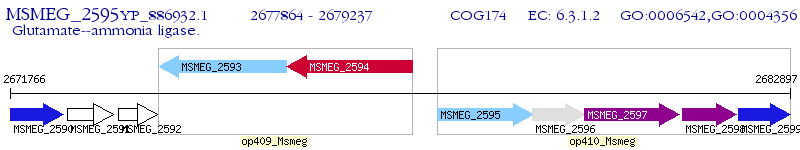
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2595 | - | - | 100% (457) | gamma-glutamylisopropylamide synthetase |
| M. smegmatis MC2 155 | MSMEG_3828 | - | 1e-92 | 42.21% (462) | glutamine synthetase |
| M. smegmatis MC2 155 | MSMEG_1116 | - | 5e-73 | 37.92% (451) | gamma-glutamylisopropylamide synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2885c | glnA4 | 0.0 | 74.50% (451) | glutamine synthetase |
| M. gilvum PYR-GCK | Mflv_4065 | - | 0.0 | 77.75% (454) | glutamate--ammonia ligase |
| M. tuberculosis H37Rv | Rv2860c | glnA4 | 0.0 | 74.50% (451) | glutamine synthetase |
| M. leprae Br4923 | MLBr_01631 | glnA2 | 5e-37 | 29.17% (408) | glutamine synthase class II |
| M. abscessus ATCC 19977 | MAB_1920 | - | 1e-40 | 31.11% (405) | glutamine synthetase |
| M. marinum M | MMAR_3294 | glnA2 | 2e-40 | 30.24% (410) | glutamine synthetase GlnA2 |
| M. avium 104 | MAV_3718 | glnA | 0.0 | 72.37% (456) | glutamine synthetase catalytic domain |
| M. thermoresistible (build 8) | TH_3396 | - | 1e-41 | 30.15% (408) | PUTATIVE glutamine synthetase, type I |
| M. ulcerans Agy99 | MUL_1335 | glnA2 | 7e-41 | 30.24% (410) | glutamine synthetase GlnA2 |
| M. vanbaalenii PYR-1 | Mvan_2278 | - | 0.0 | 84.18% (455) | glutamate--ammonia ligase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2885c|M.bovis_AF2122/97 ----------MTGPGSPPLAWTELERLVAAGD----VDTVIVAFTDMQGR
Rv2860c|M.tuberculosis_H37Rv ----------MTGPGSPPLAWTELERLVAAGD----VDTVIVAFTDMQGR
MAV_3718|M.avium_104 ---MASEDSALTGSDTAMLSLAALDRLVAAGAPETRVDTVIVAFPDMQGR
MSMEG_2595|M.smegmatis_MC2_155 -----------MSRNPGMLTQDELVQLVTDGE----IDTVIVAFCDMQGR
Mvan_2278|M.vanbaalenii_PYR-1 -----------MSTR-GMLTKAALEQLIDEGD----IDTVIVGFCDMQGR
Mflv_4065|M.gilvum_PYR-GCK MTDRTKGRLELMTTR-GMLTLEDLEKQVADGD----IDTVIVAFCDMQGR
MMAR_3294|M.marinum_M ----------------MDRQKEFVLRTLEERD----IRFVRLWFTDVLGF
MUL_1335|M.ulcerans_Agy99 ----------------MDRQKEFVLRTLEERD----IRFVRLWFTDVLGF
MLBr_01631|M.leprae_Br4923 ----------------MDRQKEFVLRTLEERD----IRFVRLWFTDVLGY
TH_3396|M.thermoresistible__bu ----------------MDRQKEFVLRSLEERD----IRFVRLWFTDVLGF
MAB_1920|M.abscessus_ATCC_1997 ----------------MDRQKEFVLRTLEERD----IRFVRLWFTDVLGY
: : : : * : * *: *
Mb2885c|M.bovis_AF2122/97 LAGKRISGRHFVDDIATRGVECCSYLLAVDVDLNTVPGYAMASWDTGYGD
Rv2860c|M.tuberculosis_H37Rv LAGKRISGRHFVDDIATRGVECCSYLLAVDVDLNTVPGYAMASWDTGYGD
MAV_3718|M.avium_104 LVGKRMDARLFVDEAAATGVECCGYLLAVDVDMNTVGGYAISGWDTGFGD
MSMEG_2595|M.smegmatis_MC2_155 LTGKRVSARLFVEEVAANGCECCNYLLAVDVDMNTVDGYAVSSWETGYGD
Mvan_2278|M.vanbaalenii_PYR-1 LTGKRVSARLFAEDVAEHGSECCNYLLAVDVDMNTVDGYAISSWETGYGD
Mflv_4065|M.gilvum_PYR-GCK LTGKRISSRLFVDDVAEHGAECCNYLLAVDVDMNTVDGYAISSWEKGYGD
MMAR_3294|M.marinum_M LKSVAIAPAELEGAFEEG----------IGFDGSSIEGFARVSES----D
MUL_1335|M.ulcerans_Agy99 LKSVAIAPAELEGAFEEG----------IGFDGSSIEGFARVSES----D
MLBr_01631|M.leprae_Br4923 LKSVAIAPAELEGAFEEG----------IGFDGSSIEGFARVSES----D
TH_3396|M.thermoresistible__bu LKSVAIAPAELEGAFEEG----------IGFDGSAIEGFARVSEA----D
MAB_1920|M.abscessus_ATCC_1997 LKSVSIAPAELEGAFEEG----------IGFDGSSIEGFARVSES----D
* . : : :..* .:: *:* . *
Mb2885c|M.bovis_AF2122/97 MVMTPDLSTLRLIPWLPG-----TALVIADLVWADGSEVAVSPRSILRRQ
Rv2860c|M.tuberculosis_H37Rv MVMTPDLSTLRLIPWLPG-----TALVIADLVWADGSEVAVSPRSILRRQ
MAV_3718|M.avium_104 LVMRPDLSTLRRIPWLPG-----TALVIADVVGADGSPVAVSPRAVLRRQ
MSMEG_2595|M.smegmatis_MC2_155 MVMTPDFSTLRRIPWLPG-----TAMVMADLSWLDGNPVAVAPRSILNHQ
Mvan_2278|M.vanbaalenii_PYR-1 MVMTPDFSTLRLLPWLPG-----SALVMADLSWTDGTPVTQAPRSILNKQ
Mflv_4065|M.gilvum_PYR-GCK MVMTPDMSTLRRVPWLPA-----TALVMADLSWTDGRPVTQAPRGILRAQ
MMAR_3294|M.marinum_M TVAHPDPSTFQVLPWVTDAGHHHSARIFCDITMPDGSPSWADPRHVLRRQ
MUL_1335|M.ulcerans_Agy99 TVAHPDPSTFQVLPWVTDAGHHHSARIFCDITMPDGSPSWADPRHVLRRQ
MLBr_01631|M.leprae_Br4923 TVAHPDPSTFQILPWATTAGHHHSARMFCDITMPDGSSSWADPRHVLRRQ
TH_3396|M.thermoresistible__bu MVARPDPSTFQVLPWTGDSGKHYSARMFCDITMPDGSPSWADPRHVLRRQ
MAB_1920|M.abscessus_ATCC_1997 TVAHPDPSTFQVLPWTTSAGKHHSARMFCDIKLPDGSPSWSDPRHVLRRQ
* ** **:: :** :* ::.*: ** ** :*. *
Mb2885c|M.bovis_AF2122/97 LDRLKARGLVADVATELEFIVFDQPYRQAWASGYRGLTPASDYNIDYAIL
Rv2860c|M.tuberculosis_H37Rv LDRLKARGLVADVATELEFIVFDQPYRQAWASGYRGLTPASDYNIDYAIL
MAV_3718|M.avium_104 LDRLAGRGLFADAATELEFMVFDEPYRQAWASGYRGLTPASDYNIDYAIS
MSMEG_2595|M.smegmatis_MC2_155 IDRLAERGLVPFVGTELEFMVFDDSFRQAWSSGYHNLTPATDYNVDYAIH
Mvan_2278|M.vanbaalenii_PYR-1 IARLAERGLVPYVGTELEFMVFDDTFRDAWKADYRGLTPATDYNVDYAMH
Mflv_4065|M.gilvum_PYR-GCK LDRLDALGLDAVAATELEFMVFDTGFRDAWAAGYRNLKPATDYNIDYAMH
MMAR_3294|M.marinum_M LTKANELGFSCYVHPEIEFFLLNPGPEDG--TEPVPIDNAGYFDQAVHVS
MUL_1335|M.ulcerans_Agy99 LIKANELGFSCYVHPEIEFFLLNPGPEDG--TEPVPIDNAGYFDQAVHVS
MLBr_01631|M.leprae_Br4923 LTKANDLGFSCYVHPEIEFFLLKAGPEDGSMPIPVPVDNAGYFDQAVHDS
TH_3396|M.thermoresistible__bu LAKAGDQGFTCYVHPEIEFFLLKPGPHDG--SPPVPADTSGYFDQSVHDT
MAB_1920|M.abscessus_ATCC_1997 LAKASDLGFSCYVHPEIEFFLLKNLPDDG--TAPIPADDAGYFDQAVHEA
: : *: . .*:**:::. :. . : ::
Mb2885c|M.bovis_AF2122/97 ASSRMEPLLRDIRLGMAGAGLRFEAVKGECNMGQQEIGFRYDEALVTCDN
Rv2860c|M.tuberculosis_H37Rv ASSRMEPLLRDIRLGMAGAGLRFEAVKGECNMGQQEIGFRYDEALVTCDN
MAV_3718|M.avium_104 ASSRMEPLLRDIRRGMAGAGLRFESVKGECNRGQQEIGFRYDEALRTCDN
MSMEG_2595|M.smegmatis_MC2_155 ASTRMEPLLRDIRRGMESAGMYCEGVKGECNLGQQEIAFRYDHARVTCDN
Mvan_2278|M.vanbaalenii_PYR-1 ASTRMEPLLRDIRLGMDGAGMYCEGVKGECNLGQQEIAFRYDHARVTCDN
Mflv_4065|M.gilvum_PYR-GCK ASTRMEPLLRDIRLGMDGAGMYCEGVKGECNLGQQEIGFRYEQALVTCDN
MMAR_3294|M.marinum_M ASK----FRRHAIDALEFMGISVEFSHHEGAPGQQEIDLRFADALSMADN
MUL_1335|M.ulcerans_Agy99 ASK----FRRHVIDALEFMGISVEFSHHEGAPGQQEIDLRFADALSMADN
MLBr_01631|M.leprae_Br4923 ALN----FRRHAIEALEFMGISVEFSHHEGAPGQQEIDLRFADALSMADN
TH_3396|M.thermoresistible__bu APN----FRRHAIEALEQMGISVEFSHHEGAPGQQEIDLRYADALSMADN
MAB_1920|M.abscessus_ATCC_1997 APN----FRRHAIDALESMGISVEFSHHEGAPGQQEIDLRYADALSMADN
* . : *. .: *: * : * ***** :*: .* .**
Mb2885c|M.bovis_AF2122/97 HAIYKNGAKEIADQHGKSLTFMAK-YDEREGNSCHIHVSLRG--TDGS--
Rv2860c|M.tuberculosis_H37Rv HAIYKNGAKEIADQHGKSLTFMAK-YDEREGNSCHIHVSLRG--TDGS--
MAV_3718|M.avium_104 HVIYKNGAKEIADQHGKSLTFMAK-YDEREGNSCHVHLSLRD--AQGG--
MSMEG_2595|M.smegmatis_MC2_155 HTIYRNGAKEIADQHGKSLTFMAK-YDEREGNSCHIHISLRGAGENGD--
Mvan_2278|M.vanbaalenii_PYR-1 HTIYRNGAKEIADQHGKSLTFMAK-YDEREGNSCHIHISLRGLDEEGRPS
Mflv_4065|M.gilvum_PYR-GCK HTIYKNGAKEIADQHGKSLTFMAK-FDESEGNSCHIHISFRGRDGSPE-D
MMAR_3294|M.marinum_M VMTFRYVIKEVALEEGVRATFMPKPFGQHAGSAMHTHMSLFEG--DVN--
MUL_1335|M.ulcerans_Agy99 VMTFRYVIKEVALEEGVRATFMPKPFGQHAGSAMHTHMSLFEG--DVN--
MLBr_01631|M.leprae_Br4923 VMTFRYVIKEVAIENGARASFMPKPFAQHPGSAMHTHMSLFEG--DVN--
TH_3396|M.thermoresistible__bu VMTFRYVVKQVAISDGVRASFMPKPFAEHPGSAMHTHMSLFEG--DTN--
MAB_1920|M.abscessus_ATCC_1997 VMTFRNVVKEVAIIDGVHATFMPKPFSDHPGSAMHTHMSLFEG--DTN--
:: *::* .* :**.* : : *.: * *:*: .
Mb2885c|M.bovis_AF2122/97 -AVFADSNGPHGMSSMFRSFVAGQLATLREFTLCYAPTINSYKRFADSSF
Rv2860c|M.tuberculosis_H37Rv -AVFADSNGPHGMSSMFRSFVAGQLATLREFTLCYAPTINSYKRFADSSF
MAV_3718|M.avium_104 -AAFADPSRPHGMSTMFCSFLAGLLATMADFTLFYAPNINSYKRFADESF
MSMEG_2595|M.smegmatis_MC2_155 -AVFADESDPHGMSPLFRSFIAGILATLRELTLFYAPNINSYKRFVDGSF
Mvan_2278|M.vanbaalenii_PYR-1 VPVFADDSDPLGMSPMFRSFIAGQLATLREMALFYAPNINSYKRFVDGSF
Mflv_4065|M.gilvum_PYR-GCK IAVFADDSDELGMSSMFRSFIAGQLATLREFTLFYAPNINSYKRFAEGSF
MMAR_3294|M.marinum_M --AFHSPDDPLQLSEVGKSFIAGILEHASEISAVTNQWVNSYKRLILGGE
MUL_1335|M.ulcerans_Agy99 --AFHSPDDPLQLSEVGKSFIAGILEHASEISAVTNQWVNSYKRLILGGE
MLBr_01631|M.leprae_Br4923 --AFHSDDDPLQLSDVGKSFIAGILEHASEISAVTNQWVNSYKRLVVGGE
TH_3396|M.thermoresistible__bu --AFHSPDDPLQLSDVAKSFIAGILEHANEISAVTNQWVNSYKRLVHGGE
MAB_1920|M.abscessus_ATCC_1997 --AFHNPDDPLQLSDVGKSFIAGILEHANEISAVTNQWVNSYKRLVQGGE
.* . . :* : **:** * ::: :*****: .
Mb2885c|M.bovis_AF2122/97 APTALAWGLDNRTCALRVVGHG----QNIRVECRVPGGDVNQYLAVAALI
Rv2860c|M.tuberculosis_H37Rv APTALAWGLDNRTCALRVVGHG----QNIRVECRVPGGDVNQYLAVAALI
MAV_3718|M.avium_104 APTALAWGLDNRTCALRVVGHG----AHTRVECRVPGGDVNPYLAVAAIV
MSMEG_2595|M.smegmatis_MC2_155 APTAVAWGFDNRTCALRVVGHG----HGMRVECRAPGGDVNQYLAVSALI
Mvan_2278|M.vanbaalenii_PYR-1 APTAVAWGLDNRTCAMRVVGHG----HGMRVECRAPGGDVNQYLAVSALI
Mflv_4065|M.gilvum_PYR-GCK APTAVAWGLDNRTCSLRVVGHG----QGMRMENRAPGGDVNQYLAVAALI
MMAR_3294|M.marinum_M APTAASWGAANRSALVRVPMYTPHKTSSRRIEVRSPDSACNPYLAFAVLL
MUL_1335|M.ulcerans_Agy99 APTAASWGAANRSALVRVPMYTPHKTSSRRIEVRSPDSACNPYLAFAVLL
MLBr_01631|M.leprae_Br4923 APTTASWGAANRSALVRVPMYTPHKTSSRRVEVRSPDSACNPYLAFAVLL
TH_3396|M.thermoresistible__bu APTAASWGAANRSALVRVPMYTPHKASSRRVEVRSPDSACNPYLTFAVLL
MAB_1920|M.abscessus_ATCC_1997 APTAASWGAANRSALVRVPMYTPNKSSSRRIEVRSPDSACNPYLTYAVLL
***: :** **:. :** : *:* * *.. * **: :.::
Mb2885c|M.bovis_AF2122/97 AGGLYGIERGLQLPEPCVGNAYQGADVER-------LPVTLADAAVLFED
Rv2860c|M.tuberculosis_H37Rv AGGLYGIERGLQLPEPCVGNAYQGADVER-------LPVTLADAAVLFED
MAV_3718|M.avium_104 AGGLYGIEQGLALPEPCAGNAYRARGVGR-------LPATLAEAAALFEH
MSMEG_2595|M.smegmatis_MC2_155 AGGLYGIERDLELPEPMAGNAYT-SGAER-------LPTTLAEAADLFAK
Mvan_2278|M.vanbaalenii_PYR-1 AGGLYGIDNELELADAMEGNAYT-SGADR-------LPTTLAEAVELFDN
Mflv_4065|M.gilvum_PYR-GCK AGGLYGIENKLELEDAVEGNAYT-SGADR-------LPTTLSEAVELFEN
MMAR_3294|M.marinum_M AAGLRGVEKGYVLGPQAEDNVWDLTPEERRTMGYRELPTSLDSALRAMES
MUL_1335|M.ulcerans_Agy99 AAGLRGVEKGYVLGPQAEDNVWDLTPEERRTMGYRELPTSLDSALRAMES
MLBr_01631|M.leprae_Br4923 AAGLRGVEKGYVLGPQAEDNVWDLTPEERLAMGYRELPTSLDSALGAMEV
TH_3396|M.thermoresistible__bu AAGLRGVEKGYVLGPEAEDNVWTLTPEERRAMGFKELPSSLGQALAEMER
MAB_1920|M.abscessus_ATCC_1997 AAGLRGVEQGYTLGPEAEDDVWGLTRAERQAMGYKELPSTLENALIAMEG
*.** *::. * .:.: * ** :* .* :
Mb2885c|M.bovis_AF2122/97 SALVREAFGEDVVAHYLNNARVELAAFNAAVTDWERIRGFERL
Rv2860c|M.tuberculosis_H37Rv SALVREAFGEDVVAHYLNNARVELAAFNAAVTDWERIRGFERL
MAV_3718|M.avium_104 SALARQVFGDDVVAHYLNNARVELAAFHAAVTDWERMRGFERL
MSMEG_2595|M.smegmatis_MC2_155 SEVARAAFGDDVVEHYLNYARVEMAAFNAAVTDWERVRGFERL
Mvan_2278|M.vanbaalenii_PYR-1 SKVARAAFGDDVVDHYLNNAAVELKAFNAAVTDWERKRGFERL
Mflv_4065|M.gilvum_PYR-GCK SKIARDAFGDDVVDHYVNNARVELKAFNAAVTDWERVRGFERL
MMAR_3294|M.marinum_M SELVAEALGEHVFDFFLRNKRREWATYRSHVTPYELSTYLSL-
MUL_1335|M.ulcerans_Agy99 SELVAEALGEHVFDFFLRNKRREWATYRSHVTPYELSTYLSL-
MLBr_01631|M.leprae_Br4923 SEFVAEALGEHVFDFFLRNKRAEWANYRSHVSLYELKTYLSL-
TH_3396|M.thermoresistible__bu SELVAEALGEHVFDFFLRNKRAEWENYRSHVTPYELKNYLSL-
MAB_1920|M.abscessus_ATCC_1997 SELVAEALGEHVFDFFLRNKRAEWARYRSNVTPFELRAYLGL-
* .. .:*:.*. .::. * :.: *: :* :