For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAHAPNRRTVVIVVFDGMKLLDVAGPAEVFAEANRFGADYVLKIASVDGADVTTSVGTRFPVDQTISDIE HADTVLVSGGDDLVGRPIDPQLVAAVRELAGRTRRLASICTGTFILATAGLLAGRRATTHWRHAGLLQRA YRDVSVEPDAIFVRDGDIYTSAGVSAGIDLALALVELDHGSELVREVARSLVVYLKRAGGQSQFSTLIES DAPQQSALRRVTEAIAADPAADHSVRSLAKVAALSTRQLTRLFQSELGTTPARFVEMVRIDAARAALDAG MSVSDTARQAGFGSPETLRRVFVTHLGVSPKAYRDRFRTAARN
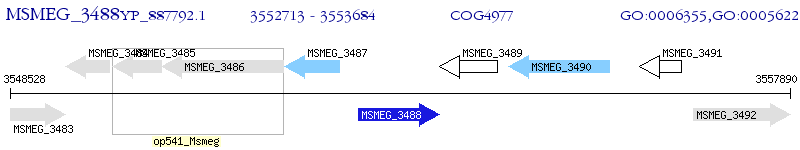
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3488 | - | - | 100% (323) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_0376 | - | 1e-77 | 47.56% (328) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_3365 | - | 2e-68 | 46.08% (319) | transcriptional regulator, AraC family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1966c | - | 3e-14 | 43.02% (86) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_1008 | - | 1e-77 | 47.43% (331) | Fis family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1931c | - | 3e-14 | 43.02% (86) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4754 | - | 1e-128 | 73.82% (317) | AraC family transcriptional regulator |
| M. marinum M | MMAR_2855 | - | 8e-66 | 44.73% (313) | transcriptional regulatory protein |
| M. avium 104 | MAV_4981 | - | 1e-129 | 74.60% (315) | transcriptional regulator, AraC family protein |
| M. thermoresistible (build 8) | TH_3120 | - | 5e-35 | 33.85% (322) | PUTATIVE transcriptional regulator, AraC family with |
| M. ulcerans Agy99 | MUL_2902 | - | 6e-65 | 44.41% (313) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_5829 | - | 1e-81 | 48.49% (332) | AraC family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3488|M.smegmatis_MC2_155 -----MAHAPNR-RTVVIVVFDGMKLLDVAGPAEVFAEANRFGADYVLKI
MAV_4981|M.avium_104 -----MADSSATPRVVVIVVFDDVTMLDVAGAGEVFAEANRFGADYLLKI
MAB_4754|M.abscessus_ATCC_1997 -----MSD----ERLITIVVFDGMKLLDLAGPAEVFAEANRFGANYRLVV
Mflv_1008|M.gilvum_PYR-GCK MTDEFRHNADPGGKTVVFALYDKVTLQDVAAPMEIFARANDFGASYRVLT
Mvan_5829|M.vanbaalenii_PYR-1 ----MAENS----RTVVFALYNKVTLQDVAAPLEIFARANDFGARYTVLL
TH_3120|M.thermoresistible__bu ---------------VAALVLDGLAIFEFGVICEVFGVDRTDDGVPPVDF
Mb1966c|M.bovis_AF2122/97 ---------------MVIVGFPG---------------------------
Rv1931c|M.tuberculosis_H37Rv ---------------MVIVGFPG---------------------------
MMAR_2855|M.marinum_M -----------MARKVVIVGFPGVQPLDVVGPFEVFAGASLLSEGGYDVA
MUL_2902|M.ulcerans_Agy99 -----------MARKVVTVGFPGVQPLDVVGPFEVFAGASLLSEGGYDVA
:.
MSMEG_3488|M.smegmatis_MC2_155 ASV---DGADVTTSVGTRFPVDQTISDI-EHADTVLVSGGD---------
MAV_4981|M.avium_104 ASV---DGRDVTTSIGTRLGVTDAVSSI-ESADTVLVAGSD---------
MAB_4754|M.abscessus_ATCC_1997 ASV---DGRDVSTSIGSPFSVTETIDSI-ECADTVLVAGGD---------
Mflv_1008|M.gilvum_PYR-GCK VSP---SGRPVGTTAFATLNVDVALDAAPDAFDTLLVPGGVPADFSFTPG
Mvan_5829|M.vanbaalenii_PYR-1 ASP---TGGAVGTTAYAQLNADLPLSEVPESIDTLLVPGGVPADFNFTPG
TH_3120|M.thermoresistible__bu TVCGPRAGQPVRTSFGATLTPDSGLDGL-AGADLVAIPAIR---------
Mb1966c|M.bovis_AF2122/97 -----------------------------DPVDTVILPGGA---------
Rv1931c|M.tuberculosis_H37Rv -----------------------------DPVDTVILPGGA---------
MMAR_2855|M.marinum_M LAS---IDERPVATGIGLEFMTTRLPDPSAPVDTVVLPGGG---------
MUL_2902|M.ulcerans_Agy99 LAS---IDERPVATGIGLEFMTTRLPDPSAPVDTVVLPGGG---------
* : :..
MSMEG_3488|M.smegmatis_MC2_155 --DLVGRPID---PQLVAAVRELAGRTRRLASICTGTFILATAGLLAGRR
MAV_4981|M.avium_104 --NLPRRPID---PELVEAVKSVADRTRRLASICTGSFILGQAGLLDGRR
MAB_4754|M.abscessus_ATCC_1997 --VLIGSPLN---PTLVAALAQLQARCRRIASICTGAFLLAQAGMLDHRR
Mflv_1008|M.gilvum_PYR-GCK LHDIPEEPTPDSVADACDMVRRLAPRARRIASVCTGSFVLAALGLLDGRR
Mvan_5829|M.vanbaalenii_PYR-1 IHDIPEEPTPDSVPDAMAMVRELAPRARRVASVCTGAFVLAALGLLDNRR
TH_3120|M.thermoresistible__bu --------QDDYLPEALDAVRAAADAGAIILTVCSGAFLAGAAGLLDGRP
Mb1966c|M.bovis_AF2122/97 -----GVDAARSEPALIDWVKAVSGTARRVVTVCTGAFLAAEAGLLG--R
Rv1931c|M.tuberculosis_H37Rv -----GVDAARSEPALIDWVKAVSGTARRVVTVCTGAFLAAEAGLLG--R
MMAR_2855|M.marinum_M -----GVDAARDNVALMDWVKAAAGTARRVVTVCTGTFLAAEAGLLDGCR
MUL_2902|M.ulcerans_Agy99 -----GVDAARDNVALMDWVKAAAGTARRVVTVCTGTFLAAEAGLLDGCR
: : ::*:*:*: . *:*
MSMEG_3488|M.smegmatis_MC2_155 ATTHWRHAGLLQRAYRDVSVEPDAIFVR-DGDIYTSAGVSAGIDLALALV
MAV_4981|M.avium_104 ATTHWHDVRLFARAFPEITVEPDALFVC-DGDVFTSAGISSGIDLALALV
MAB_4754|M.abscessus_ATCC_1997 ATTHWRHTGLLQRAFPNVTVEPDAIFVR-DANVFTSAGVSAGIDLALALV
Mflv_1008|M.gilvum_PYR-GCK ATTHWAHCQTLARQYPAVMVDPDSLFVQ-DGPFITGAGITAGIDLALAMV
Mvan_5829|M.vanbaalenii_PYR-1 ATTHWAHCQTLARTYPKVMVDPDSLFVQ-DGPFITGAGITAGIDLALALV
TH_3120|M.thermoresistible__bu CTTHWGLTEELARRYPTARVDRNVLYVD-DGDLITSAGTAAGIDACLHLV
Mb1966c|M.bovis_AF2122/97 TPSDDALG--LCRTFRPR-ISGRSGRCRPDLHAQFAEGVDRGWSHRRHRP
Rv1931c|M.tuberculosis_H37Rv TPSDDALG--LCRTFRPR-ISGRSGRCRPDLHAQFAEGVDRGWSHRRHRP
MMAR_2855|M.marinum_M ATTHWAFADRLAREFPAVDVDPDPIFIRSSPKVWTAAGVTAGIDLALALV
MUL_2902|M.ulcerans_Agy99 ATTHWAFADRLAREFPAVDVDPDPIFIRSSPKVWTAAGVTAGIDLALALV
.:. : * : :. . . * * .
MSMEG_3488|M.smegmatis_MC2_155 ELDHGSELVREVARSLVVYLKRAGGQSQFSTLIESDAPQQSALRRVTEAI
MAV_4981|M.avium_104 EMDYGTELVRAVARWLVVYLKRAGGQSQFSALVEAEPPPRSPLRKVTAAI
MAB_4754|M.abscessus_ATCC_1997 EDDHGSEMVRDVARSLVVYLKRAGGQSQFSVFVESALPTGSALRPATDAI
Mflv_1008|M.gilvum_PYR-GCK ESDFGPAVARRVARWMVVFLQRPGGQAQFSVWAESALPVTGGLRAVVDSV
Mvan_5829|M.vanbaalenii_PYR-1 ESDYGPAVARRVARWMVVFLQRPGGQAQFSVWAEATLPATGGLRAVVDSV
TH_3120|M.thermoresistible__bu RRELGSEVTNTIARHMVVPPQRDGGQRQFIDQPIPVRHSEG-FAPHLDWI
Mb1966c|M.bovis_AF2122/97 RAGTGRRRPRHRDCPDGCPLARPVSAPTRWADPVRGSGVDATRQTDLDPP
Rv1931c|M.tuberculosis_H37Rv RAGTGRRRPRHRDCPDGCPLARPVSAPTRWADPVRGSGVDATRQTDLDPP
MMAR_2855|M.marinum_M EGDHGTEIAQTVARWLVLYLRRPGGQ-TQFAAPVWMP--RAKRPSIRDVQ
MUL_2902|M.ulcerans_Agy99 EGDHGTEIAQTVARWLVLYLRRPCGQ-TQFAAPVWMP--RAKRPSIRDVQ
. * . * . .
MSMEG_3488|M.smegmatis_MC2_155 AAD-----PAADHSVRSLAKVAALSTRQLTRLFQSELGTTPARFVEMVRI
MAV_4981|M.avium_104 SAD-----PAADHSVKSLSARASLSTRQLTRLFQSELGTTPARYVEMVRI
MAB_4754|M.abscessus_ATCC_1997 AAD-----PAADHSVAKLAARAALSARQLTRLFHSELGTTPARYVELVRI
Mflv_1008|M.gilvum_PYR-GCK IAD-----PAGDHSLAAMASRAAVSERHLVRVFRDQVGMTPARFVEQARL
Mvan_5829|M.vanbaalenii_PYR-1 VAD-----PGADHSIAAMAARAAVSERHLVRMFREQVGMTPARFVEQARL
TH_3120|M.thermoresistible__bu LSN-----LDKTHTVASLARRANMSPRTFARRFVEETGRTPMQWVTDQRV
Mb1966c|M.bovis_AF2122/97 GAGGHRGRAGGAHRIGELAQRAAMSPRHFTRVFSDEVGEAPGRYVERIRT
Rv1931c|M.tuberculosis_H37Rv GAGGHRGRAGGAHRIGELAQRAAMSPRHFTRVFSDEVGEAPGRYVERIRT
MMAR_2855|M.marinum_M EA--IEAQPGGPHSIEELAHRAAMSPRHFTRVFTGEVGEAPRQYVERVRT
MUL_2902|M.ulcerans_Agy99 EA--IEAQPGGPHSIEELAHRAAMSPRHFTRVFTGEVGEAPGRYVERVRT
: * : :: * :* * :.* * : * :* ::* *
MSMEG_3488|M.smegmatis_MC2_155 DAARAALDAG-MSVSDTARQAGFGSPETLRRVFVTHLGVSPKAYRDRFRT
MAV_4981|M.avium_104 DAARAALDAG-RSVAESARLAGFGSAETLRRVFVEHLGLSPKAYRDRFRT
MAB_4754|M.abscessus_ATCC_1997 DAARGALDAG-STVTEAARIAGFGSAETLRRAFVSELGVSPKAYRERFRT
Mflv_1008|M.gilvum_PYR-GCK EAAKVLLATGDHSQDSVARRSGFGTPDTMRRTFRRNLGISPGVYRNRFRT
Mvan_5829|M.vanbaalenii_PYR-1 EAAKVLLATGDKGQDAVARRAGFGTPDTMRRTFRRNLGISPSTYRSRFRT
TH_3120|M.thermoresistible__bu LYARRLLEETDLDIHRIAARCGFGSTTLLRHHFRRVIGVTPSDYRRRFAC
Mb1966c|M.bovis_AF2122/97 EAARRQLEETHDTVVAIAARCGFGTAETMRRSFIRRVGISPDQYRKAFA-
Rv1931c|M.tuberculosis_H37Rv EAARRQLEETHDTVVAIAARCGFGTAETMRRSFIRRVGISPDQYRKAFA-
MMAR_2855|M.marinum_M EAARRQLEETEDTVVAIAARCGFGTAETMRRNFIRRVGISPDQYRKAFA-
MUL_2902|M.ulcerans_Agy99 EAARRQLEETEDTVVAIAARCGFGTAETMRRNFIRRVGISPDQYRKAFA-
*: * * .***:. :*: * :*::* ** *
MSMEG_3488|M.smegmatis_MC2_155 AARN------------
MAV_4981|M.avium_104 AARG------------
MAB_4754|M.abscessus_ATCC_1997 AAR-------------
Mflv_1008|M.gilvum_PYR-GCK TGVDA-----------
Mvan_5829|M.vanbaalenii_PYR-1 TGIDR-----------
TH_3120|M.thermoresistible__bu GSTTSGSTESGAEVTD
Mb1966c|M.bovis_AF2122/97 ----------------
Rv1931c|M.tuberculosis_H37Rv ----------------
MMAR_2855|M.marinum_M ----------------
MUL_2902|M.ulcerans_Agy99 ----------------