For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGYVSLTTLPARLKLDDMKAVAVVGFSGVQALDVVGPFETFTCASRFVDDETWQLEGYEVMLASVDGQPV PASTGLGLMACPLPDPEQVDTVVIPGGVGVRQAAADPALMTWFREAAEHARRIVSVCTGAFLVAEAGLLN GCQATTHWAFADELAQRYPEVRVDPNPMFVRSSRQVWTAAGVSAGIDLALALVEDDHSTEVAKAVARWLV LSLRRPGGQAQFAAPIWTPRARRDEIRLVQEVIEANPAAPHTIDTLATSAAMSTRHFTRVFTSEVGEGPG RYVDRVRTQAARRQLEETTDTVAAIAARCGFGTPETMRRNFIRLLGVSPEAYRKAFN
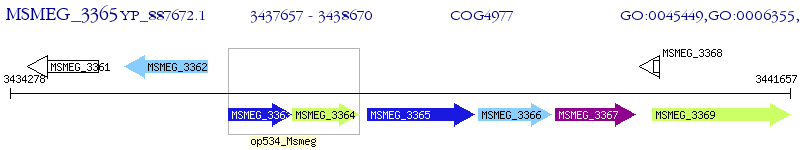
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3365 | - | - | 100% (337) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_1106 | - | e-116 | 64.35% (317) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_3488 | - | 2e-68 | 46.08% (319) | transcriptional regulator, AraC family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1966c | - | 2e-46 | 43.46% (260) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_5267 | - | 1e-114 | 64.26% (319) | AraC family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1931c | - | 2e-46 | 43.46% (260) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0942 | - | 1e-115 | 62.70% (319) | AraC family transcriptional regulator |
| M. marinum M | MMAR_2855 | - | 1e-117 | 66.77% (319) | transcriptional regulatory protein |
| M. avium 104 | MAV_4981 | - | 7e-66 | 43.57% (319) | transcriptional regulator, AraC family protein |
| M. thermoresistible (build 8) | TH_3120 | - | 2e-42 | 40.81% (272) | PUTATIVE transcriptional regulator, AraC family with |
| M. ulcerans Agy99 | MUL_2902 | - | 1e-118 | 67.08% (319) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_0972 | - | 1e-114 | 64.69% (320) | AraC family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1966c|M.bovis_AF2122/97 --------------------MVIVGFPG----------------------
Rv1931c|M.tuberculosis_H37Rv --------------------MVIVGFPG----------------------
MMAR_2855|M.marinum_M ----------------MARKVVIVGFPGVQPLDVVGPFEVFAGASLLSEG
MUL_2902|M.ulcerans_Agy99 ----------------MARKVVTVGFPGVQPLDVVGPFEVFAGASLLSEG
Mflv_5267|M.gilvum_PYR-GCK ----------------MTRTVLIVGFAGVQALDIVGPFDVFTGASLAVTA
Mvan_0972|M.vanbaalenii_PYR-1 -MHGLDAIAALANTEPMVRTVVIVGFAGVQALDVVGPFDVFTGASLSVGA
MAB_0942|M.abscessus_ATCC_1997 ----------------MRRSVVILGFPGVQALDLVGPYEVFATASRHMTG
MSMEG_3365|M.smegmatis_MC2_155 MGYVSLTTLPARLKLDDMKAVAVVGFSGVQALDVVGPFETFTCASRFVDD
MAV_4981|M.avium_104 ----------MADSSATPRVVVIVVFDDVTMLDVAGAGEVFAEANRFGAD
TH_3120|M.thermoresistible__bu --------------------VAALVLDGLAIFEFGVICEVFGVDRTDDGV
: : : .
Mb1966c|M.bovis_AF2122/97 -------------------------------------DPVDTVILPGGAG
Rv1931c|M.tuberculosis_H37Rv -------------------------------------DPVDTVILPGGAG
MMAR_2855|M.marinum_M ------GYDVALASIDERPVATGIGLEFMTTRLPDPSAPVDTVVLPGGGG
MUL_2902|M.ulcerans_Agy99 ------GYDVALASIDERPVATGIGLEFMTTRLPDPSAPVDTVVLPGGGG
Mflv_5267|M.gilvum_PYR-GCK RG--GDGYAVTVVSVGGDPVSTGTGLTLTAVPLPDPGHPIDTLVLPGGFG
Mvan_0972|M.vanbaalenii_PYR-1 RGL-GDGYDVRVVAVDGAPVSTGTGLTLAADPLPGPDEAVDTVVLPGGWG
MAB_0942|M.abscessus_ATCC_1997 TT---PGYDVKLVSADGAPVSTGSGMELVTQPLPDPEPGCDTLLLPGGIG
MSMEG_3365|M.smegmatis_MC2_155 ETWQLEGYEVMLASVDGQPVPASTGLGLMACPLPDPEQ-VDTVVIPGGVG
MAV_4981|M.avium_104 -------YLLKIASVDGRDVTTSIGTRLGVTDAVSSIESADTVLVAGSDN
TH_3120|M.thermoresistible__bu PP----VDFTVCGPRAGQPVRTSFGATLTPDSGLDGLAGADLVAIP---A
* : :.
Mb1966c|M.bovis_AF2122/97 VDAARSEPALIDWVKAVSGTARRVVTVCTGAFLAAEAGLLGRTPSDDALG
Rv1931c|M.tuberculosis_H37Rv VDAARSEPALIDWVKAVSGTARRVVTVCTGAFLAAEAGLLGRTPSDDALG
MMAR_2855|M.marinum_M VDAARDNVALMDWVKAAAGTARRVVTVCTGTFLAAEAGLLDGCRATTHWA
MUL_2902|M.ulcerans_Agy99 VDAARDNVALMDWVKAAAGTARRVVTVCTGTFLAAEAGLLDGCRATTHWA
Mflv_5267|M.gilvum_PYR-GCK ADAARRDDVLIDWITAIAPHARRVVSVCSGAVLAAQAGLLDGCVATTHWA
Mvan_0972|M.vanbaalenii_PYR-1 ADTARRDQVLIDWITAAAARARRVVSVCNGAVLAAQAGLLDGCVATTHWA
MAB_0942|M.abscessus_ATCC_1997 VSAVQRDAALMSWVRAAARGARRVVSVCNGAFLAAEAGLLDGCRATTHWA
MSMEG_3365|M.smegmatis_MC2_155 VRQAAADPALMTWFREAAEHARRIVSVCTGAFLVAEAGLLNGCQATTHWA
MAV_4981|M.avium_104 LPRRPIDPELVEAVKSVADRTRRLASICTGSFILGQAGLLDGRRATTHWH
TH_3120|M.thermoresistible__bu IRQDDYLPEALDAVRAAADAGAIILTVCSGAFLAGAAGLLDGRPCTTHWG
: . : : ::*.*:.: . ****. .
Mb1966c|M.bovis_AF2122/97 LCRTFRPRISGRSGR-----CRPDLHAQFAEGVDRGWSHRRHRPRAGTGR
Rv1931c|M.tuberculosis_H37Rv LCRTFRPRISGRSGR-----CRPDLHAQFAEGVDRGWSHRRHRPRAGTGR
MMAR_2855|M.marinum_M FADRLAREFPAVDVDPDPIFIRSSPKVWTAAGVTAGIDLALALVEGDHGT
MUL_2902|M.ulcerans_Agy99 FADRLAREFPAVDVDPDPIFIRSSPKVWTAAGVTAGIDLALALVEGDHGT
Mflv_5267|M.gilvum_PYR-GCK YAPHMAVEFPSVTVDPDPIFVRSSEKVWTAAGVTAGIDLALSLVEDDHGT
Mvan_0972|M.vanbaalenii_PYR-1 YAPQLAAEFPSVTVDPDPIFVRSSEKVWTAAGVTAGIDLALALVEDDHGT
MAB_0942|M.abscessus_ATCC_1997 IAGQLASEFPSVTVDPDPIFIRSSDKTWTAAGVTAGIDLALALVEEDHGA
MSMEG_3365|M.smegmatis_MC2_155 FADELAQRYPEVRVDPNPMFVRSSRQVWTAAGVSAGIDLALALVEDDHST
MAV_4981|M.avium_104 DVRLFARAFPEITVEPDALFVCDG-DVFTSAGISSGIDLALALVEMDYGT
TH_3120|M.thermoresistible__bu LTEELARRYPTARVDRNVLYVDDG-DLITSAGTAAGIDACLHLVRRELGS
: . . . : * * . . .
Mb1966c|M.bovis_AF2122/97 RRPRHRDCPDGCPLARPVSAPTRWADPVRGSGVDATRQTDLDPPGAGGHR
Rv1931c|M.tuberculosis_H37Rv RRPRHRDCPDGCPLARPVSAPTRWADPVRGSGVDATRQTDLDPPGAGGHR
MMAR_2855|M.marinum_M EIAQTVARWLVLYLRRPGGQ-TQFAAPVWMPRAKRPSIRDVQEAIEA---
MUL_2902|M.ulcerans_Agy99 EIAQTVARWLVLYLRRPCGQ-TQFAAPVWMPRAKRPSIRDVQEAIEA---
Mflv_5267|M.gilvum_PYR-GCK ELAQTVARHLVMYLRRPGGQ-TQFAAPVWMPRARRSPIRDVQEVIES---
Mvan_0972|M.vanbaalenii_PYR-1 ETAQIVARHLVMYLRRPGGQ-TQFAAPVWMPRARRTPIRDVQEAVEA---
MAB_0942|M.abscessus_ATCC_1997 EVAQAVARWLVLYLRRPGGQ-SQFAAPVWMPRAKRMPIRDVQEAIEA---
MSMEG_3365|M.smegmatis_MC2_155 EVAKAVARWLVLSLRRPGGQ-AQFAAPIWTPRARRDEIRLVQEVIEA---
MAV_4981|M.avium_104 ELVRAVARWLVVYLKRAGGQ-SQFSALVEAEPPPRSPLRKVTAAISA---
TH_3120|M.thermoresistible__bu EVTNTIARHMVVPPQRDGGQ-RQFIDQPIPVRHSEGFAPHLDWILSN---
. . * . :: :
Mb1966c|M.bovis_AF2122/97 GRAGGAHRIGELAQRAAMSPRHFTRVFSDEVGEAPGRYVERIRTEAARRQ
Rv1931c|M.tuberculosis_H37Rv GRAGGAHRIGELAQRAAMSPRHFTRVFSDEVGEAPGRYVERIRTEAARRQ
MMAR_2855|M.marinum_M -QPGGPHSIEELAHRAAMSPRHFTRVFTGEVGEAPRQYVERVRTEAARRQ
MUL_2902|M.ulcerans_Agy99 -QPGGPHSIEELAHRAAMSPRHFTRVFTGEVGEAPGRYVERVRTEAARRQ
Mflv_5267|M.gilvum_PYR-GCK -EPGAVHSISELARRAAMSTRHFTRAFTDEVGEAPGAYVERVRTEAARRQ
Mvan_0972|M.vanbaalenii_PYR-1 -EPGGAHSIAELARLAAMSPRHFTRVFTEEVGEAPGAYVERVRTEAARRQ
MAB_0942|M.abscessus_ATCC_1997 -EPSGSHRVPELARLAAMSPRHFTRVFTAEVGEAPSAYVERIRSEAARRQ
MSMEG_3365|M.smegmatis_MC2_155 -NPAAPHTIDTLATSAAMSTRHFTRVFTSEVGEGPGRYVDRVRTQAARRQ
MAV_4981|M.avium_104 -DPAADHSVKSLSARASLSTRQLTRLFQSELGTTPARYVEMVRIDAARAA
TH_3120|M.thermoresistible__bu --LDKTHTVASLARRANMSPRTFARRFVEETGRTPMQWVTDQRVLYARRL
* : *: * :*.* ::* * * * * :* * **
Mb1966c|M.bovis_AF2122/97 LEETHDTVVAIAARCGFGTAETMRRSFIRRVGISPDQYRKAFA-------
Rv1931c|M.tuberculosis_H37Rv LEETHDTVVAIAARCGFGTAETMRRSFIRRVGISPDQYRKAFA-------
MMAR_2855|M.marinum_M LEETEDTVVAIAARCGFGTAETMRRNFIRRVGISPDQYRKAFA-------
MUL_2902|M.ulcerans_Agy99 LEETEDTVVAIAARCGFGTAETMRRNFIRRVGISPDQYRKAFA-------
Mflv_5267|M.gilvum_PYR-GCK LEETDDTVTVIAARCGFGSAETLRRSFVRRLGVSPDQYRKTFA-------
Mvan_0972|M.vanbaalenii_PYR-1 LEETDDTVTVIAARCGFGSAETLRRNFVRRLGVSPDQYRRTFA-------
MAB_0942|M.abscessus_ATCC_1997 LEETDDTMVVIAGRCGFGSAESLRRNFVRRMGVSPDQYRKTFQRTDRSSV
MSMEG_3365|M.smegmatis_MC2_155 LEETTDTVAAIAARCGFGTPETMRRNFIRLLGVSPEAYRKAFN-------
MAV_4981|M.avium_104 LDAGR-SVAESARLAGFGSAETLRRVFVEHLGLSPKAYRDRFRTAARG--
TH_3120|M.thermoresistible__bu LEETDLDIHRIAARCGFGSTTLLRHHFRRVIGVTPSDYRRRFACGSTTSG
*: : * .***:. :*: * . :*::*. ** *
Mb1966c|M.bovis_AF2122/97 ----------
Rv1931c|M.tuberculosis_H37Rv ----------
MMAR_2855|M.marinum_M ----------
MUL_2902|M.ulcerans_Agy99 ----------
Mflv_5267|M.gilvum_PYR-GCK ----------
Mvan_0972|M.vanbaalenii_PYR-1 ----------
MAB_0942|M.abscessus_ATCC_1997 ----------
MSMEG_3365|M.smegmatis_MC2_155 ----------
MAV_4981|M.avium_104 ----------
TH_3120|M.thermoresistible__bu STESGAEVTD