For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTLAPEDRTGSPQPSVAIWLTEPSAAAVEMGALAGYDTVVLDIEHGLFDLRALDWIIPLIRAKRMRVIAK VLGPERGPIQQALDFGADAVAIPHIASADHAAQICGFAKFPPLGDRSFAGGRTSEYRGFTDDWVTAQDRN THCYPMIEDASAFDDIDAILALPVVDGIFIGPSDLSLRRDRGAYSVTDADLADIRHLARAANTAGKPWIL PAWSEAEQRLALEDGAHTIVAAMQYGAMLAGFTATMQGLQTISQDVRSAR
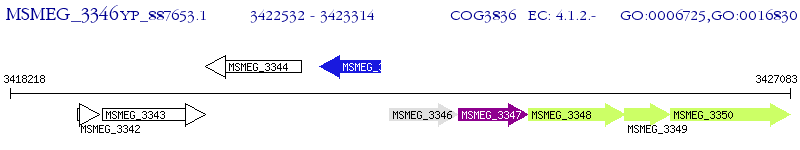
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3346 | - | - | 100% (260) | putative hydroxyacid aldolase |
| M. smegmatis MC2 155 | MSMEG_2565 | - | 6e-16 | 27.88% (226) | 4-hydroxy-2-oxovalerate aldolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4105 | - | 1e-14 | 28.79% (264) | HpcH/HpaI aldolase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0401 | - | 3e-19 | 32.24% (183) | hypothetical protein MMAR_0401 |
| M. avium 104 | MAV_5121 | - | 3e-19 | 32.28% (189) | HpcH/HpaI aldolase/citrate lyase family protein |
| M. thermoresistible (build 8) | TH_3347 | - | 5e-16 | 32.21% (149) | PUTATIVE putative HpcH/HpaI aldolase |
| M. ulcerans Agy99 | MUL_1051 | - | 1e-19 | 32.18% (202) | hypothetical protein MUL_1051 |
| M. vanbaalenii PYR-1 | Mvan_2241 | - | 2e-15 | 30.00% (190) | HpcH/HpaI aldolase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0401|M.marinum_M MKVVSNLQQRLGRGTPLWGGWITGPTWLGPEEFARAGYDYVGFDTQHGYL
MUL_1051|M.ulcerans_Agy99 MKVVSNLQQRLGRGTPLWGGWITGPTWLGPEEFARAGYDYVGFDTQHGYL
MAV_5121|M.avium_104 MTAPSNLRLALNRRTPIFGGWVTGPTLLGPEEFARAGYDYVGFDAQHGYL
Mflv_4105|M.gilvum_PYR-GCK --MTSRLQQALSSGGPVWGGWVVGPTVIGPEEYAAAGYDYVGIDVQHSYL
Mvan_2241|M.vanbaalenii_PYR-1 --MASRLQDALADNAQIWGGWVVGPTVLGPEEFAAAGYDYVGFDVQHGYL
TH_3347|M.thermoresistible__bu ------------------MALMHSRTPDVPGLAAAAGYDAVYIDLEHTAT
MSMEG_3346|M.smegmatis_MC2_155 --MTLAPEDRTGSPQPSVAIWLTEPSAAAVEMGALAGYDTVVLDIEHGLF
: : * **** * :* :*
MMAR_0401|M.marinum_M DDADVAGLLRRLEGVPIATVVRLPNADPAPIGRVLDAGADAVIIAMIESA
MUL_1051|M.ulcerans_Agy99 DDADVAGLLRRLEGVPIATVVRLPNADPGPIGRVLDAGADAVIIAMIESA
MAV_5121|M.avium_104 DDADIAGVLRRIEHLPIGTVVRLPNADAAPIGRVADAGADAVVIAMIESP
Mflv_4105|M.gilvum_PYR-GCK SDADVALMLRRLEHVPIATVVRLPDAAPAPIGRVLDAGADGVIIAMVESA
Mvan_2241|M.vanbaalenii_PYR-1 SDADVALLLRRLEHVPIATVVRLPGVDAAPIGRVLDAGADGVVIAMVESA
TH_3347|M.thermoresistible__bu PLETVAMLCAAAAGADISALVRVPSHDPSVIARVLDIGAVGVIVPHVNSK
MSMEG_3346|M.smegmatis_MC2_155 DLRALDWIIPLIRAKRMRVIAKVLGPERGPIQQALDFGADAVAIPHIASA
: : : .:.:: . . * :. * ** .* :. : *
MMAR_0401|M.marinum_M EQAAAAVAATRYPPAGVRSYGPLRA--SLGTDP-------AELQSRAGVY
MUL_1051|M.ulcerans_Agy99 EQAAAAVAATRYPPAGVRSYGPLRA--SLGTDP-------AELQSRAGVY
MAV_5121|M.avium_104 DQAAAAVAATRYPPAGVRSFGPLRV--SLGHDP-------AAHESRVSVF
Mflv_4105|M.gilvum_PYR-GCK ADAAAAVAATRYAPGGVRSFGPLRA--SLGLDP-------SALQERASVF
Mvan_2241|M.vanbaalenii_PYR-1 AQAAAAVAATRYAPGGVRSFGPLRA--GLGIEP-------AALQARASVF
TH_3347|M.thermoresistible__bu SEAESVVHSARFPPAGRRSISGPNP--VTGFRPRTAPELAAEIDRYTVVA
MSMEG_3346|M.smegmatis_MC2_155 DHAAQICGFAKFPPLGDRSFAGGRTSEYRGFTD----DWVTAQDRNTHCY
.* :::.* * ** . . * : : .
MMAR_0401|M.marinum_M AMVETAAALSTIDDICAVDGLAGIYVGPADLAISLGTGVVGALDHPAVRQ
MUL_1051|M.ulcerans_Agy99 AMVETAAAVSTIDDICAVDGPAGIYVGPADLAISLGTGVVGALDHAAVRQ
MAV_5121|M.avium_104 AMIETAAALSGITQICAVDGLTGIYVGPADLAISLGAGVIGATRQPEVLD
Mflv_4105|M.gilvum_PYR-GCK AMIETTRGLAAVDEICRVDGLTGIYVGPADLAISMGHGPTDAWSRRDVFD
Mvan_2241|M.vanbaalenii_PYR-1 AMVETARGLAAIDEICRVEGLTGIYVGPADLAISLGHGPTEAWTRPDVLN
TH_3347|M.thermoresistible__bu AMVETPQAVESVDDIAAVAGLDMILLGPSDLTAEMG--IHGDFENHRFHE
MSMEG_3346|M.smegmatis_MC2_155 PMIEDASAFDDIDAILALPVVDGIFIGPSDLSLRRDRGAYSVTDAD--LA
.*:* . .. : * : * :**:**: .
MMAR_0401|M.marinum_M AVVGIHRAASAASLHTGIHAG-AAKAGKAMAQLGFEMITLAAESQALRRG
MUL_1051|M.ulcerans_Agy99 AVVGIHRAASAANLHTGIHAG-AAKAGKAMAQLGFEMITLAAESQALRRG
MAV_5121|M.avium_104 AIVHIHKAATKAGLVTGIHAG-DGSTGHAMAQLGFDMITLAAEAQALRRG
Mflv_4105|M.gilvum_PYR-GCK VILTAQRAAAAAGVVSGIHAS-AGSAGNMLAQNGFQMITLASESQALRRG
Mvan_2241|M.vanbaalenii_PYR-1 AMQHLQRTAAKAGLVTGIHAN-TGTIGNAMAQKGFQMITLASESQALRRG
TH_3347|M.thermoresistible__bu AVGAVAEACREHGVAFGIAGIRSPDLLAHFVDLGLRFISAGTDVGMLTDA
MSMEG_3346|M.smegmatis_MC2_155 DIRHLARAANTAGKPWILPAW-SEAEQRLALEDGAHTIVAAMQYGAMLAG
: .:. . : . : * * . : : .
MMAR_0401|M.marinum_M AAEYLNEATAGTR--------
MUL_1051|M.ulcerans_Agy99 AAEYLNEATAGTR--------
MAV_5121|M.avium_104 AAEHLREATQ-----------
Mflv_4105|M.gilvum_PYR-GCK AAAHLSDARGES---GDGR--
Mvan_2241|M.vanbaalenii_PYR-1 AAAHLAEATGQSVAPGQQRYC
TH_3347|M.thermoresistible__bu ASARARELRALHRSHRT----
MSMEG_3346|M.smegmatis_MC2_155 FTATMQGLQTISQDVRSAR--
: