For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTASRLQQALADRQTVWGGWVVGPTILGPEEFAAAGYDYVGFDVQHGYLDDADVALLLRRLEHVPIATAV RLPSPDPAPIGRVLDAGADAVIIAMVETPEQAAAALAATRYAPEGVRSFGPLRASLGHDTVALQERASVI AMIETATGVAAVQDIAAVAGLTGLYVGPADLAISMGNQPTDAWTHPDVLAAFVTVRQAADAAGLAGGIHA GTGRAGKAAADMGFRMITLASESQALRRGAVEHLDEATGAGGGHEQRGYL
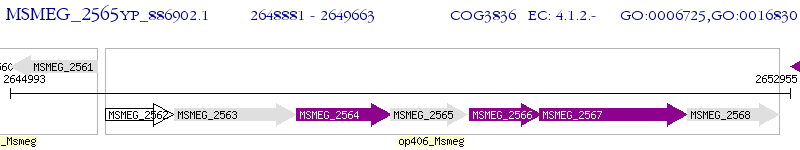
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2565 | - | - | 100% (260) | 4-hydroxy-2-oxovalerate aldolase |
| M. smegmatis MC2 155 | MSMEG_3346 | - | 6e-16 | 27.88% (226) | putative hydroxyacid aldolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4105 | - | 7e-98 | 72.29% (249) | HpcH/HpaI aldolase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0401 | - | 1e-91 | 69.39% (245) | hypothetical protein MMAR_0401 |
| M. avium 104 | MAV_5121 | - | 2e-89 | 66.53% (245) | HpcH/HpaI aldolase/citrate lyase family protein |
| M. thermoresistible (build 8) | TH_0281 | - | 9e-42 | 59.44% (143) | 4-hydroxy-2-oxovalerate aldolase |
| M. ulcerans Agy99 | MUL_1051 | - | 7e-90 | 68.57% (245) | hypothetical protein MUL_1051 |
| M. vanbaalenii PYR-1 | Mvan_2241 | - | 1e-101 | 72.37% (257) | HpcH/HpaI aldolase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0401|M.marinum_M MKVVSNLQQRLGRGTPLWGGWITGPTWLGPEEFARAGYDYVGFDTQHGYL
MUL_1051|M.ulcerans_Agy99 MKVVSNLQQRLGRGTPLWGGWITGPTWLGPEEFARAGYDYVGFDTQHGYL
MAV_5121|M.avium_104 MTAPSNLRLALNRRTPIFGGWVTGPTLLGPEEFARAGYDYVGFDAQHGYL
Mflv_4105|M.gilvum_PYR-GCK --MTSRLQQALSSGGPVWGGWVVGPTVIGPEEYAAAGYDYVGIDVQHSYL
Mvan_2241|M.vanbaalenii_PYR-1 --MASRLQDALADNAQIWGGWVVGPTVLGPEEFAAAGYDYVGFDVQHGYL
MSMEG_2565|M.smegmatis_MC2_155 -MTASRLQQALADRQTVWGGWVVGPTILGPEEFAAAGYDYVGFDVQHGYL
TH_0281|M.thermoresistible__bu --------------------------------------------------
MMAR_0401|M.marinum_M DDADVAGLLRRLEGVPIATVVRLPNADPAPIGRVLDAGADAVIIAMIESA
MUL_1051|M.ulcerans_Agy99 DDADVAGLLRRLEGVPIATVVRLPNADPGPIGRVLDAGADAVIIAMIESA
MAV_5121|M.avium_104 DDADIAGVLRRIEHLPIGTVVRLPNADAAPIGRVADAGADAVVIAMIESP
Mflv_4105|M.gilvum_PYR-GCK SDADVALMLRRLEHVPIATVVRLPDAAPAPIGRVLDAGADGVIIAMVESA
Mvan_2241|M.vanbaalenii_PYR-1 SDADVALLLRRLEHVPIATVVRLPGVDAAPIGRVLDAGADGVVIAMVESA
MSMEG_2565|M.smegmatis_MC2_155 DDADVALLLRRLEHVPIATAVRLPSPDPAPIGRVLDAGADAVIIAMVETP
TH_0281|M.thermoresistible__bu --------------------------------------------------
MMAR_0401|M.marinum_M EQAAAAVAATRYPPAGVRSYGPLRASLGTDPAELQSRAGVYAMVETAAAL
MUL_1051|M.ulcerans_Agy99 EQAAAAVAATRYPPAGVRSYGPLRASLGTDPAELQSRAGVYAMVETAAAV
MAV_5121|M.avium_104 DQAAAAVAATRYPPAGVRSFGPLRVSLGHDPAAHESRVSVFAMIETAAAL
Mflv_4105|M.gilvum_PYR-GCK ADAAAAVAATRYAPGGVRSFGPLRASLGLDPSALQERASVFAMIETTRGL
Mvan_2241|M.vanbaalenii_PYR-1 AQAAAAVAATRYAPGGVRSFGPLRAGLGIEPAALQARASVFAMVETARGL
MSMEG_2565|M.smegmatis_MC2_155 EQAAAALAATRYAPEGVRSFGPLRASLGHDTVALQERASVIAMIETATGV
TH_0281|M.thermoresistible__bu ------VAATRYPPSGVRSFGPLRSDLGHDPTVLQDRVAVFAMVETIRGV
:*****.* ****:**** .** :. : *..* **:** .:
MMAR_0401|M.marinum_M STIDDICAVDGLAGIYVGPADLAISLGTGVVGALDHPAVRQAVVGIHRAA
MUL_1051|M.ulcerans_Agy99 STIDDICAVDGPAGIYVGPADLAISLGTGVVGALDHAAVRQAVVGIHRAA
MAV_5121|M.avium_104 SGITQICAVDGLTGIYVGPADLAISLGAGVIGATRQPEVLDAIVHIHKAA
Mflv_4105|M.gilvum_PYR-GCK AAVDEICRVDGLTGIYVGPADLAISMGHGPTDAWSRRDVFDVILTAQRAA
Mvan_2241|M.vanbaalenii_PYR-1 AAIDEICRVEGLTGIYVGPADLAISLGHGPTEAWTRPDVLNAMQHLQRTA
MSMEG_2565|M.smegmatis_MC2_155 AAVQDIAAVAGLTGLYVGPADLAISMGNQPTDAWTHPDVLAAFVTVRQAA
TH_0281|M.thermoresistible__bu RNIAEICAVPGLSGIYVGPADLSLSMGHGPGGAGRVTEVDDTIDRIQQAA
: :*. * * :*:*******::*:* * * .. :::*
MMAR_0401|M.marinum_M SAASLHTGIHAGAAKAGKAMAQLGFEMITLAAESQALRRGAAEYLNEATA
MUL_1051|M.ulcerans_Agy99 SAANLHTGIHAGAAKAGKAMAQLGFEMITLAAESQALRRGAAEYLNEATA
MAV_5121|M.avium_104 TKAGLVTGIHAGDGSTGHAMAQLGFDMITLAAEAQALRRGAAEHLREATQ
Mflv_4105|M.gilvum_PYR-GCK AAAGVVSGIHASAGSAGNMLAQNGFQMITLASESQALRRGAAAHLSDARG
Mvan_2241|M.vanbaalenii_PYR-1 AKAGLVTGIHANTGTIGNAMAQKGFQMITLASESQALRRGAAAHLAEATG
MSMEG_2565|M.smegmatis_MC2_155 DAAGLAGGIHAGTGRAGKAAADMGFRMITLASESQALRRGAVEHLDEATG
TH_0281|M.thermoresistible__bu ASAGLVTGIHAGDGAAGRAMAARGFRVITLASEGQALRRGAAAHLAEATA
*.: ****. . *. * ** :****:*.*******. :* :*
MMAR_0401|M.marinum_M GTR----------------
MUL_1051|M.ulcerans_Agy99 GTR----------------
MAV_5121|M.avium_104 -------------------
Mflv_4105|M.gilvum_PYR-GCK ES---GDGR----------
Mvan_2241|M.vanbaalenii_PYR-1 QSVAPGQQRYC--------
MSMEG_2565|M.smegmatis_MC2_155 AGGGHEQRGYL--------
TH_0281|M.thermoresistible__bu PRDDLPETSTTPGSSGRYH