For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TLVVGVLISGAGCAPDGDRSATPENPVTTLPVPPNIPPFPLPDAIPVVPLPDAVPAVEMPNDIPRVQAPA DVPKMQLPSDIPPVPLPSAIPPIVFP*
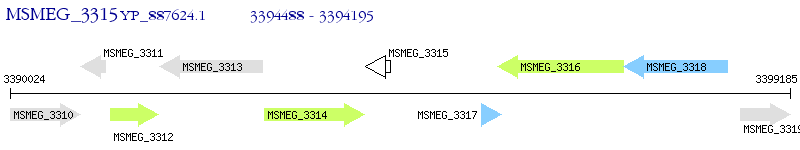
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3315 | - | - | 100% (97) | hypothetical protein MSMEG_3315 |
| M. smegmatis MC2 155 | MSMEG_1195 | - | 8e-07 | 36.11% (72) | hypothetical protein MSMEG_1195 |
| M. smegmatis MC2 155 | MSMEG_5700 | - | 5e-06 | 34.12% (85) | secreted protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1470c | - | 4e-05 | 32.32% (99) | proline, glycine, valine-rich secreted protein |
| M. gilvum PYR-GCK | Mflv_1992 | - | 2e-16 | 47.89% (71) | hypothetical protein Mflv_1992 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00568 | - | 4e-05 | 33.85% (65) | hypothetical protein MLBr_00568 |
| M. abscessus ATCC 19977 | MAB_4537c | - | 2e-06 | 36.84% (95) | hypothetical protein MAB_4537c |
| M. marinum M | MMAR_0601 | - | 5e-08 | 34.41% (93) | hypothetical protein MMAR_0601 |
| M. avium 104 | MAV_4824 | - | 5e-07 | 32.95% (88) | hypothetical protein MAV_4824 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3723 | rpfE | 3e-06 | 40.58% (69) | resuscitation-promoting factor RpfE |
| M. vanbaalenii PYR-1 | Mvan_1015 | - | 4e-06 | 34.52% (84) | hypothetical protein Mvan_1015 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3315|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1992|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0601|M.marinum_M MFSSAATVVGCKTLGGVLDVVLGVSMAAADVRMALVEGEDGD-GVIVEED
MAV_4824|M.avium_104 -----------------MDTVLGVSMAPTAVRMVLVEGENGD-GATVDED
Mvan_1015|M.vanbaalenii_PYR-1 -------------------------MTASSIRWVLVDGATGEESVTIDRG
MUL_3723|M.ulcerans_Agy99 --------------------------------------------------
Mb1470c|M.bovis_AF2122/97 --------------------------------------------------
MLBr_00568|M.leprae_Br4923 --------------------------------------------------
MAB_4537c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_3315|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1992|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0601|M.marinum_M SFDLDR--NPVTSSASDQVVAAILGTREGAADAGLRLSSIGVTWTDPVQ-
MAV_4824|M.avium_104 NFDVATSEDAATVTASDQVLSAILGTRQGAAEGGYQLASTGVTFTDPVE-
Mvan_1015|M.vanbaalenii_PYR-1 SVPVDD------AFQADALLDRLLGR---------PVHAIGLTWTVEAER
MUL_3723|M.ulcerans_Agy99 --------------------------------------------------
Mb1470c|M.bovis_AF2122/97 --------------------------------------------------
MLBr_00568|M.leprae_Br4923 --------------------------------------------------
MAB_4537c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_3315|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1992|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0601|M.marinum_M -AAALRDALAWHKVENVMLVSGFLAAAALGQAVGGALSYARTAVLFIEPD
MAV_4824|M.avium_104 -AAALRDKLAAHKVENVMLVSAFLAAAALAQAVGHRTNYAHTALLYIEPD
Mvan_1015|M.vanbaalenii_PYR-1 TASAVWQGLTDRGVENVIAVSDVESAEALACGIADLAGCERVVVGLAEPG
MUL_3723|M.ulcerans_Agy99 --------------------------------------------------
Mb1470c|M.bovis_AF2122/97 --------------------------------------------------
MLBr_00568|M.leprae_Br4923 --------------------------------------------------
MAB_4537c|M.abscessus_ATCC_199 -MLVLTRWFRRALVSGVCVAGLLGAGVVPALADPGDPPDPGIEEPGPPPP
MSMEG_3315|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1992|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0601|M.marinum_M TATLAVVDTADGSVSEVCQQLLAEDDDAAVAEVVAMVAAAESLPARPDGV
MAV_4824|M.avium_104 TATLAVVDSADGSIADVRRQPLPEDDEAAVAELASMVSNAENLETRPDAV
Mvan_1015|M.vanbaalenii_PYR-1 TSVIAAVTPDG-----------------VTADRVDPAALTDVDGFDPDAI
MUL_3723|M.ulcerans_Agy99 --------------------------------------------------
Mb1470c|M.bovis_AF2122/97 -----------------------------------------------MTL
MLBr_00568|M.leprae_Br4923 --------------------------------------------------
MAB_4537c|M.abscessus_ATCC_199 PPLPWELPPPPPPPAPDAPPPAEVPPVPAPVPTTPPPGAPPTGDVPPLPV
MSMEG_3315|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1992|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0601|M.marinum_M YVVGTGVDVAMIKPALDAATSLTVSVSEEQEMALARGAALACGNAPLFAA
MAV_4824|M.avium_104 FVVGSGIDIPLIKPALEAATTLPLTAPEEPDTALARGAALASAHAPLFAS
Mvan_1015|M.vanbaalenii_PYR-1 FVLGSG-DVAGVVSALEQRSTVPVVSADEADLALARGAALASASAVSILD
MUL_3723|M.ulcerans_Agy99 --MKNVRKTLIVAAITETFVTVPTAATANADDGLDPNAVGADTTGFDPNL
Mb1470c|M.bovis_AF2122/97 MAIVNRFNIKVIAGAGLFAAAIALSPDAAADPLMTGGYACIQGMAGDAPV
MLBr_00568|M.leprae_Br4923 MKVFDRFNTAILVSAGLCGAALAFSPSAAAAPLSMPTGGPTCIEQMAG--
MAB_4537c|M.abscessus_ATCC_199 GLLKNSPNNGDVVGVAQPITIVFAAPVTDKKAAEAAVKITTSKPAPGYFY
MSMEG_3315|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1992|M.gilvum_PYR-GCK ----------------------------------------------MARA
MMAR_0601|M.marinum_M TTAALAYAQDLNDPDPLGESDLGDELSPDQEPAGRRSGLLIGSGLAVVAT
MAV_4824|M.avium_104 STAALAYAQ---DPGTGAINPLAVAPGYFETPADAGADGLAYSAVPDDPD
Mvan_1015|M.vanbaalenii_PYR-1 AQAAPARRR-------------------------LSPTATLASVLATAVV
MUL_3723|M.ulcerans_Agy99 PAAPGAFEP-----------------------PAPQDAPPAPELADADPD
Mb1470c|M.bovis_AF2122/97 AAG-----------------------------------DPVAAGGPAATG
MLBr_00568|M.leprae_Br4923 ---------------------------------------LGAVPAAVPAV
MAB_4537c|M.abscessus_ATCC_199 WYTDQQLRWKP-------------TQFWPANTDVNVNAGGTKWSFKVGDA
MSMEG_3315|M.smegmatis_MC2_155 -----MTLVVG--VLISGAG------------------------------
Mflv_1992|M.gilvum_PYR-GCK CPLVIIAAVLGLTVLAAGGGR-----------------------------
MMAR_0601|M.marinum_M AAVVALEVALAIDIRPTVALQPRPNENHLVAPPPQAAPPPQVAAPEPKIE
MAV_4824|M.avium_104 EFFTGTQAAATELVDADYADR---SSFKLVGSAVAAIFVIGVVALVVSLA
Mvan_1015|M.vanbaalenii_PYR-1 TFVVSLSVALGMTLTPDSPAP----QMARAVTEPTPEPVEKPVVKPAPVK
MUL_3723|M.ulcerans_Agy99 LPPPAPDAMIAPVDMPDGPAP-----------------------------
Mb1470c|M.bovis_AF2122/97 ACSAALTDMAGVPFVAPGPVP-----------------------------
MLBr_00568|M.leprae_Br4923 LPGPVAGAVPSVPVVPSGGFP----------------------------P
MAB_4537c|M.abscessus_ATCC_199 FVSTVDDATYTMTVTRNGVVER------------------------TMPI
MSMEG_3315|M.smegmatis_MC2_155 --CAPDGDRS--------ATPENPVTTLPVPPNIPPFPLPDAIPVVPLPD
Mflv_1992|M.gilvum_PYR-GCK AAAAPGQDDHGAPVIPPPAFPALPLPVLSIPAGLPPFPYPADVPVVQLPA
MMAR_0601|M.marinum_M LARPRVAPRAASPQLPAPAAPVQRAPEVPV-LPAPAVPEPVVAVPPLVPI
MAV_4824|M.avium_104 IAIRPTANVRPSPNQNVVVAPTRPAPAAPAPAPAPEAPAPAPAAAPQAPA
Mvan_1015|M.vanbaalenii_PYR-1 AAPPEPEAAPPPPAAPPVAEPVAVEIPAPAAPALPEAPPPVYDEPLAPPP
MUL_3723|M.ulcerans_Agy99 EDAPPAPDAFAPPAPEDVPPAPEPVAFEPPAPDVAPPPAPKVYTVNWDAI
Mb1470c|M.bovis_AF2122/97 AAAPVPIGAPVPIPGAPVPIPGAPVPIPGAPVPIPGGPVPIPGAPVPVPA
MLBr_00568|M.leprae_Br4923 VVPPGGFPPVVPPGGFLPVVPPGGFPPVVPPGGFPPVVPPGVAPAVGPLL
MAB_4537c|M.abscessus_ATCC_199 SMGKHDKKHETKNGTYYVSEKFQKMVMDSSTYGVPVNSAEGYKLDVYWAT
.
MSMEG_3315|M.smegmatis_MC2_155 AVPAV-EMPN--------DIPRVQAPAD--VPKMQLPSDIPPVPLPSAIP
Mflv_1992|M.gilvum_PYR-GCK DIPTV-EFPS--------AVPPVTLPGP--VPSVQLPAAIPPIQLPAPIP
MMAR_0601|M.marinum_M VVPPV-RVPSGTDTFLRPSVPKVQAPVPRQQPKAPAPRVEVPASPPPVVS
MAV_4824|M.avium_104 PAPEA-PAPAPAAP---APAPVQVAPVP--VPEAPAPAPAAPPPPPPAVA
Mvan_1015|M.vanbaalenii_PYR-1 VVAPA-PAYVPPPPPANYLPPAPAAPAY--VPPAQVPAYVPPAQQPAYVP
MUL_3723|M.ulcerans_Agy99 AQCEP-GGNWGINTGNGYSGGLQFTASTWRASGGAGSAAGASREEQIRVA
Mb1470c|M.bovis_AF2122/97 VPAPV-IPVGTPLIALGPVLAGAPGDGVVSAPIIGMSGVKDALTDPAPAG
MLBr_00568|M.leprae_Br4923 AEDSA-ALGSGILAGKGVPTASPQAALSGLVVLPGPPASVEAPPAAAPVV
MAB_4537c|M.abscessus_ATCC_199 RLSNSGIFVHAAPWSVGAQGKSDTSHGCINVNTDNATWFFNQSHPGDPVI
.
MSMEG_3315|M.smegmatis_MC2_155 PIVFP---------------------------------------------
Mflv_1992|M.gilvum_PYR-GCK RGLL----------------------------------------------
MMAR_0601|M.marinum_M PNPRGRGGLRLPGLGRIPNPGRGPAGGSLGPGRRGHFGGGAPGRSGPSGG
MAV_4824|M.avium_104 PIP-----VPIP----LPIPGLG------GPG----FGG-PPGHGGGDGG
Mvan_1015|M.vanbaalenii_PYR-1 PAPQ-------------------------QPRLRDRIIERIPIINRFHEP
MUL_3723|M.ulcerans_Agy99 ENVLR-----------------------------------SQGIGAWPVC
Mb1470c|M.bovis_AF2122/97 GPVPG--------------------------------QPVLPGPSASAPA
MLBr_00568|M.leprae_Br4923 AAATP-----------------------------------LLCCGEAAPP
MAB_4537c|M.abscessus_ATCC_199 VKNSP--------------------------------GGPYKDYDGYDDW
MSMEG_3315|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1992|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0601|M.marinum_M GHGGQFGGGGHGGRFGGGGHGGGFGGGRGGGGHGGGFGGFGGGHGGGGHG
MAV_4824|M.avium_104 GHG----GGGFPGFPGGGGHGG------------GGFPGFGGGHGGFGHH
Mvan_1015|M.vanbaalenii_PYR-1 EY------------------------------------------------
MUL_3723|M.ulcerans_Agy99 GRRG----------------------------------------------
Mb1470c|M.bovis_AF2122/97 GAR-----------------------------------------------
MLBr_00568|M.leprae_Br4923 VR------------------------------------------------
MAB_4537c|M.abscessus_ATCC_199 QRF-----------------------------------------------
MSMEG_3315|M.smegmatis_MC2_155 ------------------
Mflv_1992|M.gilvum_PYR-GCK ------------------
MMAR_0601|M.marinum_M GGFGGGHGGGFGGGHGGR
MAV_4824|M.avium_104 ------------------
Mvan_1015|M.vanbaalenii_PYR-1 ------------------
MUL_3723|M.ulcerans_Agy99 ------------------
Mb1470c|M.bovis_AF2122/97 ------------------
MLBr_00568|M.leprae_Br4923 ------------------
MAB_4537c|M.abscessus_ATCC_199 ------------------