For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSLVSEVGALAEFLRSHRQRLTPEQVGIQTQGARRVPGLRREEVAMRAGISVEYYIRLEQGRERSPSLAV CEALASALVLGRYETSHLYELAEPRKSCPDERLGRPRVPEASRVLLDTLETPALIQNRYADVVATNPAGE ALSPNIRVGVNRIRALFTDPAERALHLDWDRVTENAVAQLRLAVGGSVGRSPARELITELLHTSTRFRNL WSHHEVAHAPVSPVRLQHPAVGRLVLFGQQLLIAGTDDLSMVVYHADPAASSWSGLQRLLGLASEAEPNA
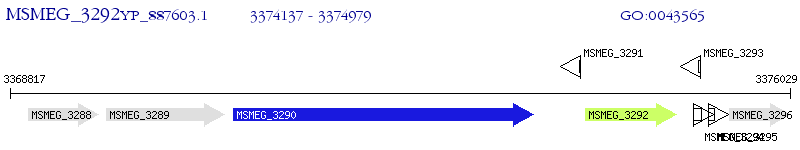
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3292 | - | - | 100% (280) | DNA-binding protein |
| M. smegmatis MC2 155 | MSMEG_0523 | - | 1e-43 | 41.26% (269) | DNA-binding protein |
| M. smegmatis MC2 155 | MSMEG_0260 | - | 2e-42 | 39.48% (271) | DNA-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_5170 | - | 6e-26 | 32.97% (273) | XRE family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4641 | - | 5e-29 | 31.97% (269) | putative DNA-binding protein |
| M. marinum M | MMAR_3947 | - | 7e-07 | 28.65% (192) | transcriptional regulatory protein |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5465 | - | 3e-38 | 39.29% (280) | XRE family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5170|M.gilvum_PYR-GCK -MDTRAQRRLALGEFLRARREAIRRADVGLPELPRARTG-GLRREEVAVT
MAB_4641|M.abscessus_ATCC_1997 -MAGTDERRRELGAFLRVRREHLVRADYGLPPVGRSRTT-GLRREEIAYL
MSMEG_3292|M.smegmatis_MC2_155 -MSLVSEVG-ALAEFLRSHRQRLTPEQVGIQTQG-ARRVPGLRREEVAMR
Mvan_5465|M.vanbaalenii_PYR-1 MMANVADANRELADFLKRARSAVDPARAGLPADGRIRRVPGLRREEVALL
MMAR_3947|M.marinum_M -MPNR-------GGFVHAARERAG-----------------LTCRELAEK
* . *:: *. * .*:*
Mflv_5170|M.gilvum_PYR-GCK AGVSVTWYTWLEQGRDINPSKQVLDAVADALRLSPAEHEYVLALGGYA--
MAB_4641|M.abscessus_ATCC_1997 SGVSVTWYTWLEQGRDINPSRQVLEAIALEMRLSAAERTYLLELAGFAPL
MSMEG_3292|M.smegmatis_MC2_155 AGISVEYYIRLEQGRERSPSLAVCEALASALVLGRYETSHLYELAEPRKS
Mvan_5465|M.vanbaalenii_PYR-1 AGVSTDYYARLEQGRRITPSAAVLDAIARALDLGDTGRAHLGRLVGASPT
MMAR_3947|M.marinum_M AGCSLRYVHDIEQG--CSPSIRVLNGLVRALQLSAWEQRFLFAACGRILP
:* * : :*** .** * :.:. : *. .:
Mflv_5170|M.gilvum_PYR-GCK -VDAAVSRGPLPPHIQHFLDALEGYPAFAITPDWGIAAWNTTYAALYPNV
MAB_4641|M.abscessus_ATCC_1997 PHSATPDPAPVPDHLLRLVENLVPSPAFVVSADWSIGGWNRAYELLYPGI
MSMEG_3292|M.smegmatis_MC2_155 CPDERLGRPRVPEASRVLLDTL-ETPALIQNRYADVVATNPAGEALSPNI
Mvan_5465|M.vanbaalenii_PYR-1 RRRSSRPVQRVRPGLYQLLDSLDETPAMILGRRTDVLATNRAARVLFADF
MMAR_3947|M.marinum_M DPALTDT-----DISQRYLDALQPHPAAALHPSWRFKAWNQQWQRLFQGV
:: * ** . . * * ..
Mflv_5170|M.gilvum_PYR-GCK ERVPETDRNLLWLVFTDPYIHELLPNWDTDSRRFLAEFRADAGPRVGEPS
MAB_4641|M.abscessus_ATCC_1997 ASIVPADRNLLRFIFTDPYVRQMLPDWPITSRRFLAEYRAEAGALLGDPR
MSMEG_3292|M.smegmatis_MC2_155 RVG----VNRIRALFTDPAERALHLDWDRVTENAVAQLRLAVGGSVGRSP
Mvan_5465|M.vanbaalenii_PYR-1 DAMPPRERNYARWMFTSEQARNLFADWSVQARTAVENLRLDFGGDPHDPA
MMAR_3947|M.marinum_M LLAP----SMHQWLYLSTTSRRILRNWDEVSQWCVGWLRYGLAVDVD--A
. :: . : : :* :. : * .
Mflv_5170|M.gilvum_PYR-GCK YTALVSRLSSESPHFAAAWQATSIERFTSRERLFRHPAAGELRLEHHQVA
MAB_4641|M.abscessus_ATCC_1997 HRALVDRLQQQSPEFARAWAEHEVGRFTSRERRFHHPEAGELIFEHHRLS
MSMEG_3292|M.smegmatis_MC2_155 ARELITELLHTSTRFRNLWSHHEVAHAPVSPVRLQHPAVGRLVLFGQQLL
Mvan_5465|M.vanbaalenii_PYR-1 VIELVDELSAASAEFRMWWAEHRVFQRTYGTKRLRHPVVGDLSVDYETFT
MMAR_3947|M.marinum_M VKPMVSALMP-VTEFRREWEAQVIPVDPATRLWVVRDGGTELRIDMRFWH
:: * ..* * : . . : * . .
Mflv_5170|M.gilvum_PYR-GCK PVDVPDIQIVAYLPVPDSVASERLRMLTGQADTAR---
MAB_4641|M.abscessus_ATCC_1997 PSDLPDLHIVIYLPLDGTNTRTKLEALLSGGR------
MSMEG_3292|M.smegmatis_MC2_155 IAGTDDLSMVVYHADPAASSWSGLQRLLGLASEAEPNA
Mvan_5465|M.vanbaalenii_PYR-1 MPGDAEQTLFVYLAEAGSPSRDALNLLLSWSANSV---
MMAR_3947|M.marinum_M ASPMPGGLLLGVILDAG---------------------
:.