For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDLGDYLRSRRALVTPADVGLPAAGHRRVPGLRREEVALLAGLSVDYYSRLEQGRERSPSAQVLDALAA VLRLDEDGRRHLFAVAGLTPRARLAPVPDRVDPALMRLMATWPDNPALVYNRAYDLLASNALADALYDGI LHHNMLMMVFTEPRARELYADWPVIAADAVAGFRMNHGIAPHDPRINAVLTELLDRSAEFTELWSRHDAR GKSATVKSFRHPQVGAVTLQMQTFDVRSAPGQELVVYQAEPGTPSADALKLLGSLAATAEA
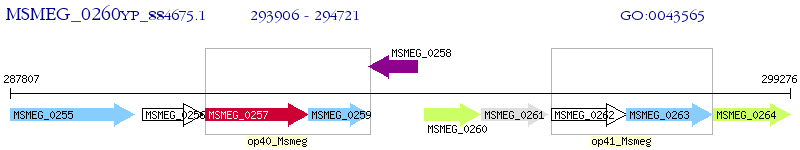
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0260 | - | - | 100% (271) | DNA-binding protein |
| M. smegmatis MC2 155 | MSMEG_0523 | - | 4e-57 | 45.96% (272) | DNA-binding protein |
| M. smegmatis MC2 155 | MSMEG_1626 | - | 4e-51 | 43.07% (267) | putative DNA-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_5170 | - | 2e-34 | 32.47% (271) | XRE family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4641 | - | 4e-40 | 34.59% (266) | putative DNA-binding protein |
| M. marinum M | MMAR_3947 | - | 5e-07 | 26.82% (179) | transcriptional regulatory protein |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_1958 | - | 7e-05 | 29.17% (120) | transcriptional regulator, XRE family protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5465 | - | 2e-54 | 44.16% (274) | XRE family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5170|M.gilvum_PYR-GCK -MDTRAQRRLALGEFLRARREAIRRADVGLPELP-RARTGGLRREEVAVT
MAB_4641|M.abscessus_ATCC_1997 -MAGTDERRRELGAFLRVRREHLVRADYGLPPVG-RSRTTGLRREEIAYL
MSMEG_0260|M.smegmatis_MC2_155 -MT-------DLGDYLRSRRALVTPADVGLPAAG-HRRVPGLRREEVALL
Mvan_5465|M.vanbaalenii_PYR-1 MMANVADANRELADFLKRARSAVDPARAGLPADGRIRRVPGLRREEVALL
MMAR_3947|M.marinum_M ---------------MPNRGGFVHAA----------RERAGLTCRELAEK
TH_1958|M.thermoresistible__bu -----VESRPASGHDAEVTRTAAPAFGAMLRDWR---RRRRLSQLDLALE
. * ::*
Mflv_5170|M.gilvum_PYR-GCK AGVSVTWYTWLEQGRDINPSKQVLDAVADALRLSPAEHEYVLALGGYA--
MAB_4641|M.abscessus_ATCC_1997 SGVSVTWYTWLEQGRDINPSRQVLEAIALEMRLSAAERTYLLELAGFAPL
MSMEG_0260|M.smegmatis_MC2_155 AGLSVDYYSRLEQGRERSPSAQVLDALAAVLRLDEDGRRHLFAVAGLTPR
Mvan_5465|M.vanbaalenii_PYR-1 AGVSTDYYARLEQGRRITPSAAVLDAIARALDLGDTGRAHLGRLVGASPT
MMAR_3947|M.marinum_M AGCSLRYVHDIEQG--CSPSIRVLNGLVRALQLSAWEQRFLFAACGRI--
TH_1958|M.thermoresistible__bu AGVSARHVSFMETGR-AAPSRTMVLRLAEVLEVPPREQNGLLVAAGYAPV
:* * :* * ** :: :. : : : : *
Mflv_5170|M.gilvum_PYR-GCK -VD---AAVSRGPLPPHIQHFLDALEGYPAFAITPDWGIAAWNTTYAALY
MAB_4641|M.abscessus_ATCC_1997 PHS---ATPDPAPVPDHLLRLVENLVPSPAFVVSADWSIGGWNRAYELLY
MSMEG_0260|M.smegmatis_MC2_155 ARL---APVP-DRVDPALMRLMATWPDNPALVYNRAYDLLASNALADALY
Mvan_5465|M.vanbaalenii_PYR-1 RRR---SSRPVQRVRPGLYQLLDSLDETPAMILGRRTDVLATNRAARVLF
MMAR_3947|M.marinum_M ------LPDPALTDTDISQRYLDALQPHPAAALHPSWRFKAWNQQWQRLF
TH_1958|M.thermoresistible__bu YAERDLDDPEMAAVRHGVQRVLDAHDPYPCVAVDRGWRVVQTNAGAAVLL
: : *. . * *
Mflv_5170|M.gilvum_PYR-GCK PNVERVPETDRNLLWLVFTDPYIHELLPNWDTDSRRFLAEFRADAGPRVG
MAB_4641|M.abscessus_ATCC_1997 PGIASIVPADRNLLRFIFTDPYVRQMLPDWPITSRRFLAEYRAEAGALLG
MSMEG_0260|M.smegmatis_MC2_155 DG-----ILHHNMLMMVFTEPRARELYADWPVIAADAVAGFRMNHGIAPH
Mvan_5465|M.vanbaalenii_PYR-1 ADFDAMPPRERNYARWMFTSEQARNLFADWSVQARTAVENLRLDFGGDPH
MMAR_3947|M.marinum_M QG----VLLAPSMHQWLYLSTTSRRILRNWDEVSQWCVGWLR--YGLAVD
TH_1958|M.thermoresistible__bu DGVADHLLEQPNALRIALHPDGLAPRIRNLAQWRHHLISRLRREVALS-R
. . : : * .
Mflv_5170|M.gilvum_PYR-GCK EPSYTALVSRLSSESPHFA---------AAWQATSIERFTSRERLFRHPA
MAB_4641|M.abscessus_ATCC_1997 DPRHRALVDRLQQQSPEFA---------RAWAEHEVGRFTSRERRFHHPE
MSMEG_0260|M.smegmatis_MC2_155 DPRINAVLTELLDRSAEFT---------ELWSRHDARGKSATVKSFRHPQ
Mvan_5465|M.vanbaalenii_PYR-1 DPAVIELVDELSAASAEFR---------MWWAEHRVFQRTYGTKRLRHPV
MMAR_3947|M.marinum_M VDAVKPMVSALMP-VTEFR---------REWEAQVIPVDPATRLWVVRDG
TH_1958|M.thermoresistible__bu APGAVALLAEIESYPGGFDDPGDLGGVVVPLELYTTSRRLLSFLSTVTTF
:: : *
Mflv_5170|M.gilvum_PYR-GCK AGELRLEHHQVAPVDVPDIQIVAYLPVPDSVASERLRMLTGQADTAR---
MAB_4641|M.abscessus_ATCC_1997 AGELIFEHHRLSPSDLPDLHIVIYLPLDGTNTRTKLEALLSGGR------
MSMEG_0260|M.smegmatis_MC2_155 VGAVTLQMQTFDVRSAPGQELVVYQAEPGTPSADALKLLGSLAATAEA--
Mvan_5465|M.vanbaalenii_PYR-1 VGDLSVDYETFTMPGDAEQTLFVYLAEAGSPSRDALNLLLSWSANSV---
MMAR_3947|M.marinum_M GTELRIDMRFWHASPMPGGLLLGVILDAG---------------------
TH_1958|M.thermoresistible__bu GTALDLTAAELSIEAFLPADERTASALAGPTSPAAASFPDPDTGTAAARQ
: . .
Mflv_5170|M.gilvum_PYR-GCK -
MAB_4641|M.abscessus_ATCC_1997 -
MSMEG_0260|M.smegmatis_MC2_155 -
Mvan_5465|M.vanbaalenii_PYR-1 -
MMAR_3947|M.marinum_M -
TH_1958|M.thermoresistible__bu P