For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLYSALPDAQGLYDPQNEADSCGVAMIADIQGRKSHRIVSDGLIALENLEHRGAAGAEASSGDGAGILIQ LPVELLREVVDFELPAPTADGDNTFAAGVCFLPQDPVARSAARERIEAIAVEEGLEVLGWRKLPVDPDGA GIGATALGCMPYMAQFFVTAPETGGARPGGVDLDRLVYPVRKRAEREDVYFPTLSSRTMVYKGMLTTMQL PQYFLDLRDERCQSAIAIVHSRFSTNTFPSWPLAHPFRFVAHNGEINTVRGNRNRMHAREAKLASPLIPG DLSRLSPICSPEASDSASFDEVLELLHLGGRSLPHAVLMMIPEAWENATTMGAAERAFWQFHASLMEPWD GPACVTFTDGTVVGAVLDRNGLRPGRWWRTVDDRVILASEAGVLDVPSGEIVAKGRLQPGKMLLIDTAAG RIVPDDEIKAQLSAAEPYEEWLHAGLLDLKTLPERGVAKPNHDSVVRRQIAFGYTEEELRVLLTPMAASG NEPLGSMGTDTPTAVLSSRSRLLYDYFVELFAQVTNPPLDAIREEVVTSMARVMGPEENLLEPTAASCRQ IVLRAPVIDNDDLGKIVHINDDGEHPGLRATVLRALYDVERGGEGLAEALDDLRRRASEAIADGKRILVL SDRDSDHTRAPIPSLLAVSAVHHHLVRTKERTKVALVVESGDAREVHHIALLIGFGAAAVNPYLAFESIE DLIREGELTGIEPAAAVRNYIKALCKGVVKVMSKMGISTVASYTAAQVFEAIGLDRGVIDEYFTGTTSQL GGVGLDVLADEVKMRHRRAYPENPTERVHRRLEVGGEYAFRREGELHLFTPEVVFLLQHSTRTGRRDIFA KYSEEVDRLSRKGGTLRGLFELKTGVRPPVPLEEVEPVESIVTRFNTGAMSYGSISAEAHETMAIAMNKL GGRSNSGEGGEDVDRLYDPRRRSAVKQVASGRFGVTSDYLVNATDIQIKMAQGAKPGEGGQLPGYKVYPN IAKTRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANSDARIHVKLVSSVGVGTVAAGVSKAHADVVL ISGHDGGTGAAPLTSLKHAGAPWEIGLADTQQTLVLNGLRDRITVQCDGGLRTARDVVVAMLLGAEEYGF ATAPLVVSGCIMMRVCHLDTCPVGVATQNPELRARFNGKPEFVENFFTFIAEDIRRYLAELGFRSIDEAV GHAELLDTAEGVAHWKTRGLDLDPIFAVPTDAHGAKLTQRRKLREQDHGLDQALDRTLIQLAEGALEDAH PVRLELPVRNVNRTVGTLLGSEVTRRYGAAGLPDDTIHVTLTGSAGQSIGAFLPPGITLELVGDANDYVG KGLSGGRVVVRPPDDVLFLPEDNVIAGNTLMYGATSGEVYLRGRVGERFCARNSGALAVVEGVGDHACEY MTGGRVVILGKTGRNMAAGMSGGIAFVLGLDPARVNTEMVQLQRLDPEDLEWLHHVVTDHVRYTGSTLGT SLLSDWPRRSAQFTKIMPTDYQRVLQATRMAKAEGRDVDSAIMEASRG
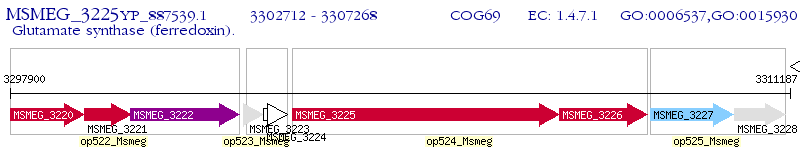
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3225 | - | - | 100% (1518) | ferredoxin-dependent glutamate synthase 1 |
| M. smegmatis MC2 155 | MSMEG_6459 | - | 0.0 | 63.62% (1542) | ferredoxin-dependent glutamate synthase 1 |
| M. smegmatis MC2 155 | MSMEG_5594 | - | 1e-35 | 33.66% (309) | ferredoxin-dependent glutamate synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3889c | gltB | 0.0 | 64.18% (1541) | putative ferredoxin-dependent glutamate synthase |
| M. gilvum PYR-GCK | Mflv_3593 | - | 0.0 | 86.55% (1517) | glutamate synthase (ferredoxin) |
| M. tuberculosis H37Rv | Rv3859c | gltB | 0.0 | 64.18% (1541) | ferredoxin-dependent glutamate synthase |
| M. leprae Br4923 | MLBr_00061 | gltB | 0.0 | 62.13% (1537) | putative ferredoxin-dependent glutamate synthase |
| M. abscessus ATCC 19977 | MAB_0091 | - | 0.0 | 62.44% (1544) | putative ferredoxin-dependent glutamate synthase |
| M. marinum M | MMAR_5413 | gltB | 0.0 | 63.21% (1541) | ferredoxin-dependent glutamate synthase |
| M. avium 104 | MAV_0166 | - | 0.0 | 63.21% (1541) | ferredoxin-dependent glutamate synthase 1 |
| M. thermoresistible (build 8) | TH_2102 | gltB | 0.0 | 64.01% (1545) | PROBABLE FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE |
| M. ulcerans Agy99 | MUL_5037 | gltB | 0.0 | 62.95% (1544) | ferredoxin-dependent glutamate synthase |
| M. vanbaalenii PYR-1 | Mvan_2822 | - | 0.0 | 86.30% (1518) | glutamate synthase (ferredoxin) |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3889c|M.bovis_AF2122/97 ------MTPKRVGLYNPAFEHDSCGVAMVVDMHGRRSRDIVDKAITALLN
Rv3859c|M.tuberculosis_H37Rv ------MTPKRVGLYNPAFEHDSCGVAMVVDMHGRRSRDIVDKAITALLN
MLBr_00061|M.leprae_Br4923 ------MTAKRVGLYNPAFEHDSCGVAMVVDMYGRRTRDIVDKAITALLN
MMAR_5413|M.marinum_M ------MTPKRVGLYNPAFEHDSCGVAMVVDMHGRRSRDIVDKAITALLN
MUL_5037|M.ulcerans_Agy99 ------MTPKRVGLYNPAFEHDSCGVAMVVDMHGRRSRDIVDKAITALLN
MAV_0166|M.avium_104 ----MHMTPKRAGLYNPAFEHDACGVAMVVDMHGRRSRDIVDKAITALLN
TH_2102|M.thermoresistible__bu ------MTPKAQGLYNPAFEHDACGVAMVADMYGRRSRDIVDKAITALLN
MAB_0091|M.abscessus_ATCC_1997 ------MPHPATGLYNPAYEHDSCGVAMVVDMHGRRSRDIVDKAITALLN
Mflv_3593|M.gilvum_PYR-GCK MSTLFSALPQPQGLYDPANESDSCGVAMVVDIQGRSSHSIVTDGLIALEH
Mvan_2822|M.vanbaalenii_PYR-1 --MLFSALPDAQGLYDPGNESDSCGVAMVTDIQGRRSHSIVADGLTALEH
MSMEG_3225|M.smegmatis_MC2_155 --MLYSALPDAQGLYDPQNEADSCGVAMIADIQGRKSHRIVSDGLIALEN
***:* * *:*****:.*: ** :: ** ..: ** :
Mb3889c|M.bovis_AF2122/97 LEHRGAQGAEPRSGDGAGILIQVPDEFLREAV----DFELPAP-----GS
Rv3859c|M.tuberculosis_H37Rv LEHRGAQGAEPRSGDGAGILIQVPDEFLREAV----DFELPAP-----GS
MLBr_00061|M.leprae_Br4923 LEHRGAQGAEPRSGDGAGILIQVPDAFLRAVV----DFELPAA-----SS
MMAR_5413|M.marinum_M LEHRGAAGAEPRSGDGAGILIQVPDAFLREVV----DFELPAP-----GS
MUL_5037|M.ulcerans_Agy99 LEHRGAAGAEPRSGDGAGILIQVPDAFLREVV----DFELPAP-----GS
MAV_0166|M.avium_104 LEHRGAQGAEPRSGDGAGILIQVPDAFLREVV----DFELPPE-----GS
TH_2102|M.thermoresistible__bu LEHRGAQGAEPNTGDGAGILLQVPDEFLRAVVEEQGDFELPPP-----GS
MAB_0091|M.abscessus_ATCC_1997 LEHRGAAGAEPNSGDGAGIMLQIPDKFFRAVLAEQGSFELPAE-----GS
Mflv_3593|M.gilvum_PYR-GCK LEHRGAAGAEPNSGDGAGILIQLPVDLLRDVA----DFDLPPATGDG-NT
Mvan_2822|M.vanbaalenii_PYR-1 LEHRGAAGAEPTSGDGAGILIQLPVDLLRAVT----DFALPDPTGDGANT
MSMEG_3225|M.smegmatis_MC2_155 LEHRGAAGAEASSGDGAGILIQLPVELLREVV----DFELPAPTADGDNT
****** ***. :******::*:* ::* . .* ** .:
Mb3889c|M.bovis_AF2122/97 YATGIAFLPQSSKDAAAACAAVQKIAEAEGLQVLGWRSVPTD--DSSLGA
Rv3859c|M.tuberculosis_H37Rv YATGIAFLPQSSKDAAAACAAVQKIAEAEGLQVLGWRSVPTD--DSSLGA
MLBr_00061|M.leprae_Br4923 YATGIAFLPQSSKDAAAACAAVEKIAVAEGLHVLGWRNVPTD--ASSLGA
MMAR_5413|M.marinum_M YATGIAFLPQSSKDAAAACAAVEKIAEAEGLQVLGWRNVPTD--DSSLGA
MUL_5037|M.ulcerans_Agy99 YATGIAFLPQSSKDAAAACAAVEKIGEAEGLQVLGWRNVPTD--DSSLGA
MAV_0166|M.avium_104 YATGIAFLPQSSKDAATACAAVEKIAEAEGLTVLGWRNVPTD--DSSLGA
TH_2102|M.thermoresistible__bu YATGIAFLPQSAKDAAQACDAVEKIVEAEGLTVLGWREVPID--DSSLGA
MAB_0091|M.abscessus_ATCC_1997 YASGIAFLPQGSKDAATACEAVEKIVEAEGLTVLGWREVPHD--DSSLGA
Mflv_3593|M.gilvum_PYR-GCK FAAGICYLPQDPVQRDAARAAVETIAAEENLDVLGWREVPVDPAGAEIGR
Mvan_2822|M.vanbaalenii_PYR-1 FAAGICFLPQDLAARESARGTVEAIAAEEGLEVLGWREMPVDPDGADLGA
MSMEG_3225|M.smegmatis_MC2_155 FAAGVCFLPQDPVARSAARERIEAIAVEEGLEVLGWRKLPVDPDGAGIGA
:*:*:.:***. * :: * *.* *****.:* * : :*
Mb3889c|M.bovis_AF2122/97 LSRDAMPTFRQVFLAG-------ASGMALERRCYVVRKRAEHELGTKGPG
Rv3859c|M.tuberculosis_H37Rv LSRDAMPTFRQVFLAG-------ASGMALERRCYVVRKRAEHELGTKGPG
MLBr_00061|M.leprae_Br4923 LSRDAMPTFRQVFMAG-------ATGLTLERRAYVIRKRAEHELGTKGPG
MMAR_5413|M.marinum_M LSRDAMPTFRQVFLAG-------ASDMTLERRCYVIRKRAEHELGTKGPG
MUL_5037|M.ulcerans_Agy99 LSRDAMPTFRQVFLAG-------ASDMALERRCYVIRKRAEHELGTKGPG
MAV_0166|M.avium_104 LSRDAMPTFRQVFMAG-------ASGMTLERRAYLVRKRAEHELGTKGPG
TH_2102|M.thermoresistible__bu LARDAMPTFRQLFLGG-------ATGIDLERRAYVVRKRAEHELGTKGPG
MAB_0091|M.abscessus_ATCC_1997 LARDAMPTFRQLFIAG-------ASGIDLERRVYVVRKRIEHELGNQGSG
Mflv_3593|M.gilvum_PYR-GCK TALDCMPHMAQLFVAAPERDGKRPGGIELDRRVYPLRKRAE---------
Mvan_2822|M.vanbaalenii_PYR-1 TALSCMPHMAQLFVAAPELNGARPGGIDLDRRVYPLRKRAE---------
MSMEG_3225|M.smegmatis_MC2_155 TALGCMPYMAQFFVTAPETGGARPGGVDLDRLVYPVRKRAE---------
: ..** : *.*: . . .: *:* * :*** *
Mb3889c|M.bovis_AF2122/97 QDGPGRETVYFPSLSGQTLVYKGMLTTPQLKAFYLDLQDERLTSALGIVH
Rv3859c|M.tuberculosis_H37Rv QDGPGRETVYFPSLSGQTLVYKGMLTTPQLKAFYLDLQDERLTSALGIVH
MLBr_00061|M.leprae_Br4923 QDGPGRETIYFPSLSSQTFVYKGMLTTPQLKAFYRDLQDERLTSALGIVH
MMAR_5413|M.marinum_M QDGPGRETVYFPSLSGRTFVYKGMLTTPQLKAFYLDLQDDRLTSALGIVH
MUL_5037|M.ulcerans_Agy99 QDGPGRETVYFPSLSGRTFVYKGMLTTPQLKAFYLDLQDDRLTSALGIVH
MAV_0166|M.avium_104 QDGPGRETVYFPSLSGQTFVYKGMLTTPQLKAFYLDLQDDRLTSALGIVH
TH_2102|M.thermoresistible__bu QDGPGRETVYFPSLSGQTFVYKGMLTTPQLKAFYLDLQDERLTSALAIVH
MAB_0091|M.abscessus_ATCC_1997 RGSLGEETVYFPSLSGRTFVYKGMLTTPQLRAFYLDLQDERVESALGIVH
Mflv_3593|M.gilvum_PYR-GCK -----QTGVYFPSLSSRTMVYKGMLTTMQLPQYFPDLRDERCISAIAIVH
Mvan_2822|M.vanbaalenii_PYR-1 -----RAGVYFPSLSSRTMVYKGMLTTMQLPQYFSDLRDERCISAIAIVH
MSMEG_3225|M.smegmatis_MC2_155 -----REDVYFPTLSSRTMVYKGMLTTMQLPQYFLDLRDERCQSAIAIVH
. :***:**.:*:******** ** :: **:*:* **:.***
Mb3889c|M.bovis_AF2122/97 SRFSTNTFPSWPLAHPFRRIAHNGEINTVTGNENWMRAREALIKTDIFGS
Rv3859c|M.tuberculosis_H37Rv SRFSTNTFPSWPLAHPFRRIAHNGEINTVTGNENWMRAREALIKTDIFGS
MLBr_00061|M.leprae_Br4923 SRFSTNTFPSWPLAHPFRRIAHNGEINTVTGNENWMRAREALMKTDIFGS
MMAR_5413|M.marinum_M SRFSTNTFPSWPLAHPFRRIAHNGEINTVTGNENWMRAREALIKTDVFGT
MUL_5037|M.ulcerans_Agy99 SRFSTNTFPSWPLAHPFRRIAHNGEINTVTGNENWMRAREALIKTDVFGT
MAV_0166|M.avium_104 SRFSTNTFPSWPLAHPFRRIAHNGEINTVTGNENWMRAREALIKTDVFGT
TH_2102|M.thermoresistible__bu SRFSTNTFPSWPLAHPFRRIAHNGEINTVTGNENWMRAREALIRTDLFG-
MAB_0091|M.abscessus_ATCC_1997 SRFSTNTFPSWPLAHPYRRVAHNGEINTVAGNENWMRAREALIKTDVFGD
Mflv_3593|M.gilvum_PYR-GCK SRFSTNTFPSWPLAHPFRFVAHNGEINTVRGNRNRMHAREAMLASAHIP-
Mvan_2822|M.vanbaalenii_PYR-1 SRFSTNTFPSWPLAHPFRFVAHNGEINTVRGNRNRMHAREAMLASAKIP-
MSMEG_3225|M.smegmatis_MC2_155 SRFSTNTFPSWPLAHPFRFVAHNGEINTVRGNRNRMHAREAKLASPLIP-
****************:* :********* **.* *:**** : : :
Mb3889c|M.bovis_AF2122/97 AADVEKLFPICTPGASDTARFDEVLELLHLGGRSLAHAVLMMIPEAWERH
Rv3859c|M.tuberculosis_H37Rv AADVEKLFPICTPGASDTARFDEVLELLHLGGRSLAHAVLMMIPEAWERH
MLBr_00061|M.leprae_Br4923 KADMEKLFPICTPGASDTARFDEALELLHLGGRSLVHAVLMMIPEAWERH
MMAR_5413|M.marinum_M EANVEKLFPICTPGASDTARFDEVLELLHLGGRSLAHAVLMMIPEAWERH
MUL_5037|M.ulcerans_Agy99 EANVEKLFPICTPGASDTARFDEVLELLHLGGRSLAHAVLMMIPEAWERH
MAV_0166|M.avium_104 EADVEKLFPICTPGASDTARFDEVLELLHLGGRSLAHAVLMMIPEAWERH
TH_2102|M.thermoresistible__bu GQDLEKVAPICTPGASDTARFDEVLELLHLGGRSLPHAVLMMIPEAWEQH
MAB_0091|M.abscessus_ATCC_1997 PAQLEKIFPICTRGASDTARFDEALELLHLGGRPLHHAVLMMIPEAWERH
Mflv_3593|M.gilvum_PYR-GCK -GDLSRLSPICTPDASDSASFDEVLELLHLGGRSLPHAVLMMIPEAWENS
Mvan_2822|M.vanbaalenii_PYR-1 -GDLTRLSPICTPDASDSASFDEVLELLHLGGRSLPHAVLMMIPEAWENN
MSMEG_3225|M.smegmatis_MC2_155 -GDLSRLSPICSPEASDSASFDEVLELLHLGGRSLPHAVLMMIPEAWENA
:: :: ***: ***:* ***.*********.* ************.
Mb3889c|M.bovis_AF2122/97 ESMDPARRAFYQYHASLMEPWDGPASMTFTDGTVVGAVLDRNGLRPSRIW
Rv3859c|M.tuberculosis_H37Rv ESMDPARRAFYQYHASLMEPWDGPASMTFTDGTVVGAVLDRNGLRPSRIW
MLBr_00061|M.leprae_Br4923 ESMDPARRAFYQYHASLMEPWDGPASMTFTDGVVVGAVLDRNGLRPSRIW
MMAR_5413|M.marinum_M ASMDPSRRAFYQYHSSLMEPWDGPASMTFTDGTIIGAVLDRNGLRPSRIW
MUL_5037|M.ulcerans_Agy99 ASMDPSRRAFYQYHSSLMEPWDGPASMTFTDGTMVGAVLDRNGLRPSRIW
MAV_0166|M.avium_104 ESMDPARRAFYQYHASLMEPWDGPASMTFTDGTVIGAVLDRNGLRPSRIW
TH_2102|M.thermoresistible__bu ESMDPARRAFYEFHDSLMEPWDGPASVCFTDGTVIGAVLDRNGLRPSRIW
MAB_0091|M.abscessus_ATCC_1997 ENMSAELRSFYEFHASLMEPWDGPASVCFTDGTIVGAVLDRNGLRPSRVW
Mflv_3593|M.gilvum_PYR-GCK TTMEPHERAFWQFHASLMEPWDGPACVTFTDGTLVGAVLDRNGLRPGRWW
Mvan_2822|M.vanbaalenii_PYR-1 TTMGDAERAFWQFHASLMEPWDGPACVTFTDGTLVGAVLDRNGLRPGRWW
MSMEG_3225|M.smegmatis_MC2_155 TTMGAAERAFWQFHASLMEPWDGPACVTFTDGTVVGAVLDRNGLRPGRWW
.* *:*:::* **********.: ****.::***********.* *
Mb3889c|M.bovis_AF2122/97 VTDDGLVVMASEAGVLDLHPSTVVRRMRLQPGRMFLVDTAQGRIVSDEEI
Rv3859c|M.tuberculosis_H37Rv VTDDGLVVMASEAGVLDLHPSTVVRRMRLQPGRMFLVDTAQGRIVSDEEI
MLBr_00061|M.leprae_Br4923 ITDAGLVVMASEAGVLDLDPSIVVRRMRLQPGRMFLVDTAQGRIVSDEEI
MMAR_5413|M.marinum_M VTEDGLVVMASEAGVLDLDPSTVVQRMRLQPGRMFLVDTAQGRIVSDEEI
MUL_5037|M.ulcerans_Agy99 VTEDGLVVMASEAGVLDLDPSTVVQRMRLQPGRMFLVDTAQGRIVSDEAI
MAV_0166|M.avium_104 VTEDGLVVMASEAGVLDLDPATVVRRMRLQPGRMFLVDTAQGRIVSDEEI
TH_2102|M.thermoresistible__bu VTADGLVVMASEAGVLNLDPATVVRKMRLQPGRMFLVDTAQGRIIDDEEI
MAB_0091|M.abscessus_ATCC_1997 VTNDGLVVMASEAGVLDLDPSTVVQRTRLQPGRMFLVDTTQGRIVSDEEV
Mflv_3593|M.gilvum_PYR-GCK RTLDDRLILASESGVLDVPSSEIVAKGRLQPGKMLLVDTAAGRIVSDDEI
Mvan_2822|M.vanbaalenii_PYR-1 RTIDDRIILASETGVLDVPSAEIVAKGRLQPGKMFLVDTAAGRIVSDDEV
MSMEG_3225|M.smegmatis_MC2_155 RTVDDRVILASEAGVLDVPSGEIVAKGRLQPGKMLLIDTAAGRIVPDDEI
* . :::***:***:: .. :* : *****:*:*:**: ***: *: :
Mb3889c|M.bovis_AF2122/97 KADLAAEHPYQEWLDNGLVPLDELPEGKDVRMPHHRIVMRQLAFGYTYEE
Rv3859c|M.tuberculosis_H37Rv KADLAAEHPYQEWLDNGLVPLDELPEGKDVRMPHHRIVMRQLAFGYTYEE
MLBr_00061|M.leprae_Br4923 KAKLAAEHPYQEWLDKGLVPIDELPAGKYVRMPHHRLVMRQLAFGYTYEE
MMAR_5413|M.marinum_M KAELAAEHPYQEWLDNNLVPLEKLPSGEYARMAHERVVLRQLVFGYTYEE
MUL_5037|M.ulcerans_Agy99 KAELAAEHPYQEWLDNNLVPLEKLPSGEYARMAHERVVLRQLVFGYTYEE
MAV_0166|M.avium_104 KAELAAEHPYQEWLDRNLVPLDDLPQGDYKRMPHDRLVKRQQTFGYTYEE
TH_2102|M.thermoresistible__bu KAELAAEHPYQEWLDAGLFPLDELPQGDYQRMPHHRVVLRQRVFGYTSEE
MAB_0091|M.abscessus_ATCC_1997 KAELAAAEPYQRWLEEGLVRLEQLPDRPHQHMPHNRIVLRQQVFGYTYEE
Mflv_3593|M.gilvum_PYR-GCK KQELSAAQPYGEWLHSGLLELTTLPDRGHIQPNHESVVRRQVSFGYTEEE
Mvan_2822|M.vanbaalenii_PYR-1 KEKLAHAEPYGEWLHSGLLELNALPDRARAQPNHESVVRRQISFGYTEED
MSMEG_3225|M.smegmatis_MC2_155 KAQLSAAEPYEEWLHAGLLDLKTLPERGVAKPNHDSVVRRQIAFGYTEEE
* .*: .** .**. .*. : ** : *. :* ** **** *:
Mb3889c|M.bovis_AF2122/97 LNLLVAPMARLGAEPIGSMGTDTPVAVLSQRPRMLYDYFHQLFAQVTNPP
Rv3859c|M.tuberculosis_H37Rv LNLLVAPMARLGAEPIGSMGTDTPVAVLSQRPRMLYDYFHQLFAQVTNPP
MLBr_00061|M.leprae_Br4923 LNLLVAPMVRAGSEPIGSMGADTPIAVLSRRPRMLYDYFHQLFAQVTNPP
MMAR_5413|M.marinum_M LNLLVAPMVRTGAEPIGSMGTDTPIAVLSRRPRMLYDYFQQLFAQVTNPP
MUL_5037|M.ulcerans_Agy99 LNLLVAPMVRTGAEPIGSMGTDTPIAVLSRRPRMLYDYFQQLFAQVTNPP
MAV_0166|M.avium_104 LNLLVAPMVRTGAEPIGSMGTDTPVAVLSQRPRMLYDYFQQLFAQVTNPP
TH_2102|M.thermoresistible__bu LNLIVGPMARTGAEPLGSMGTDTPIAVLSQRSRLLFDYFQQMFAQVTNPP
MAB_0091|M.abscessus_ATCC_1997 INLLVAPMARTGAEALGSMGTDTPIAVLSNRSRMLFDYFQQLFAQVTNPP
Mflv_3593|M.gilvum_PYR-GCK LRILLAPMAASGAEPLGSMGTDTPAAVLSQRSKLLYDYFVELFAQVTNPP
Mvan_2822|M.vanbaalenii_PYR-1 LRILLTPMAASGVEPLGSMGTDTPPAVLSQRSKLLYDYFVELFAQVTNPP
MSMEG_3225|M.smegmatis_MC2_155 LRVLLTPMAASGNEPLGSMGTDTPTAVLSSRSRLLYDYFVELFAQVTNPP
:.::: **. * *.:****:*** **** *.::*:*** ::********
Mb3889c|M.bovis_AF2122/97 LDAIREEVVTSLQGTTGGERDLLNPDENSCHQIVLPQPILRNHELAKLVS
Rv3859c|M.tuberculosis_H37Rv LDAIREEVVTSLQGTTGGERDLLNPDQNSCHQIVLPQPILRNHELAKLVS
MLBr_00061|M.leprae_Br4923 LDAIREEVVTSLQGTTGGEYDLLSPDENSCHQIWLPQPILRNYELAKLVN
MMAR_5413|M.marinum_M LDAIREEVVTSLQGTTGGERDLLNPDENSCHQIQLPQPILRNHELAKLIN
MUL_5037|M.ulcerans_Agy99 LDAIREEVVTSLQGTTGGERDLLNPDENSCHQIQLPQPILRNHELAKLIN
MAV_0166|M.avium_104 LDAIREEVVTSLQGTTGGERDLLTPTELSCHQIVLSQPILRNRELAKLVN
TH_2102|M.thermoresistible__bu LDAIREEVITSLQGVIGPEGDLLNPGPESCRQIVLPQPILRNHELAKLIN
MAB_0091|M.abscessus_ATCC_1997 LDAIREEVVTSLGGVIGPEGDLLHPTAESCHQILLPQPVLHNDELDKLIH
Mflv_3593|M.gilvum_PYR-GCK LDAIREEIVTSMARVMGPEQNLLEPGAASCRQIKLPWPVLDNDELGKIVH
Mvan_2822|M.vanbaalenii_PYR-1 LDAIREEVVTSMARIMGPEQNLLEPSAASCRQIKLTWPVLDNDELNKIVH
MSMEG_3225|M.smegmatis_MC2_155 LDAIREEVVTSMARVMGPEENLLEPTAASCRQIVLRAPVIDNDDLGKIVH
*******::**: * * :** * **:** * *:: * :* *::
Mb3889c|M.bovis_AF2122/97 LDPNDKVNGRPHGLRSKVIRCLYRVSEGGAGLAAALEEVRGAAAAAIADG
Rv3859c|M.tuberculosis_H37Rv LDPNDKVNGRPHGLRSKVIRCLYRVSEGGAGLAAALEEVRGAAAAAIADG
MLBr_00061|M.leprae_Br4923 LDPADEVNGRPHGMRSTVVSCLYPVADAGAGLAAALTEVRAQASAAIVGG
MMAR_5413|M.marinum_M LDPETVINGRPHGLKSKVVRCLYPVAEGGAGLAAALNEVRAQASAAIADG
MUL_5037|M.ulcerans_Agy99 LDPETVINGRPHGLKSKVVRCLYPVAEGGSGLAAALNEVRAQASAAIADG
MAV_0166|M.avium_104 LNPDDEVNGRPHGMRSKVIRCLYPVAEGGAGLAAALEDVRAQASAAIADG
TH_2102|M.thermoresistible__bu VDPDHEVRGRKHGMRAAVVRCLFPVNRGGQGLKEALDRVRAKVSAAIREG
MAB_0091|M.abscessus_ATCC_1997 LDPADTVNGRAHGFSSRVIRCLYPVAEGGAGLRTALESVRAEVSAAIAGG
Mflv_3593|M.gilvum_PYR-GCK INDD----GEHPGLRTAVLRALYDVERGGEGLADALEDLRLRACEAIAKG
Mvan_2822|M.vanbaalenii_PYR-1 INDD----GEQPGLRTTVLRALYNVERGGEGLAEAIEDLQLRASDAIAKG
MSMEG_3225|M.smegmatis_MC2_155 INDD----GEHPGLRATVLRALYDVERGGEGLAEALDDLRRRASEAIADG
:: *. *: : *: .*: * .* ** *: :: .. ** *
Mb3889c|M.bovis_AF2122/97 ARIIILSDRESDEEMAPIPSLLAVAGVHHHLVRERTRTQVGLVVESGDAR
Rv3859c|M.tuberculosis_H37Rv ARIIILSDRESDEEMAPIPSLLAVAGVHHHLVRERTRTQVGLVVESGDAR
MLBr_00061|M.leprae_Br4923 ARVIILSDRNSDEQMAPIPSLLSVAAVHHHLVRDRTRTHVGLVVESGDAR
MMAR_5413|M.marinum_M ARIIVLSDRESDERMAPIPALLAVAGVHHHLVRERSRTKVGLVVESGDAR
MUL_5037|M.ulcerans_Agy99 ARIIVLSDRESDERMAPIPALLAVAGVHHHLVRERTRTKVGLVVESGDAR
MAV_0166|M.avium_104 ARVIILSDRESDEKMAPIPSLLAVAGVHHHLVRDRTRTHVGLVVESGDAR
TH_2102|M.thermoresistible__bu ARIIVLSDRESDEQLAPIPSLLSVAAVHHHLVRDRTRTQVGLVVEAGDAR
MAB_0091|M.abscessus_ATCC_1997 AQVIILSDRESDDLMAPIPSLLAVAAVHHHLVRERSRTKVGLVVEAGDAR
Mflv_3593|M.gilvum_PYR-GCK ARTLVISDRDSDHTKAPVPSLLAVSAVHHHLVGTKQRTSVALVVESGDAR
Mvan_2822|M.vanbaalenii_PYR-1 AHTLVISDRDSDHTKAPVPSLLAVSAVHHHLVRTKKRTTVALVVESGDAR
MSMEG_3225|M.smegmatis_MC2_155 KRILVLSDRDSDHTRAPIPSLLAVSAVHHHLVRTKERTKVALVVESGDAR
: :::***:**. **:*:**:*:.****** : ** *.****:****
Mb3889c|M.bovis_AF2122/97 EVHHMAALVGFGAAAINPYLVFESIEDMLDRGVIEGIDRTAALNNYIKAA
Rv3859c|M.tuberculosis_H37Rv EVHHMAALVGFGAAAINPYLVFESIEDMLDRGVIEGIDRTAALNNYIKAA
MLBr_00061|M.leprae_Br4923 EVHHMAALIGFGAAAVNPYLVFESIEEMLDRGVIDGIDRKTALNNYIKAT
MMAR_5413|M.marinum_M EVHHMAMLLGCGAAAVNPYLAFESIEDMLDRGVIDGIDRNTALSNYVKAA
MUL_5037|M.ulcerans_Agy99 EVHHMAMLLGCGAAAVNPYLAFASIEDMLDRGVIDGIDRNTALSNYVKAA
MAV_0166|M.avium_104 EVHHMAALVGFGAAAINPYMVFESIEDMLDRGVIEGIDRNTALNNYVKAA
TH_2102|M.thermoresistible__bu EVHHMAALIGFGASAINPYLAFESIEDMLDRNLLEGLDRETALNNYIKAA
MAB_0091|M.abscessus_ATCC_1997 EVHHVAALVGFGAAAVNPYMAFESIEDLIDRGVITGVDRDKAIRNYIKAA
Mflv_3593|M.gilvum_PYR-GCK EVHHIAMLIGFGAAAVNPYLAFESIEDLIREGELTGIEPSTAVRNYLKAL
Mvan_2822|M.vanbaalenii_PYR-1 EVHHIAMLIGFGAAAVNPYLAFESIEDLIREGELTGVDAATAVRNYLKAL
MSMEG_3225|M.smegmatis_MC2_155 EVHHIALLIGFGAAAVNPYLAFESIEDLIREGELTGIEPAAAVRNYIKAL
****:* *:* **:*:***:.* ***::: .. : *:: *: **:**
Mb3889c|M.bovis_AF2122/97 GKGVLKVMSKMGISTLASYTGAQLFQAVGISEQVLDEYFTGLTCPTGGIT
Rv3859c|M.tuberculosis_H37Rv GKGVLKVMSKMGISTLASYTGAQLFQAVGISEQVLDEYFTGLTCPTGGIT
MLBr_00061|M.leprae_Br4923 GQGVLKIMSKMGISTLASYVGAQLFQAVGISEDVLDEYFSGLSCPTGGIT
MMAR_5413|M.marinum_M GKGVLKVMSKMGISTLASYTGAQLFQAVGVSEEVLNEYFSGLTCATGGIT
MUL_5037|M.ulcerans_Agy99 GKGVLKVMSKMGISTLASYTGAQLFQAVGVSEEVLNEYFSGLTCATGGIT
MAV_0166|M.avium_104 GKGVLKVMSKMGISTLASYTGAQLFQAVGISEDVLDEYFTGLSCPIGGIT
TH_2102|M.thermoresistible__bu GKGVLKVMSKMGISTLRSYTGAQLFQAIGISQEVLDEYFTGLSCPTGGID
MAB_0091|M.abscessus_ATCC_1997 GKGVLKVMSKMGISTLASYTGAQLFQAIGLSQELLDEYFTGLACPTGGIG
Mflv_3593|M.gilvum_PYR-GCK GKGVMKVMSKMGISTVASYTAAQAFEAIGIDRDVIDEYFTGTPTQLGGIG
Mvan_2822|M.vanbaalenii_PYR-1 GKGVMKVMSKMGISTVASYTAAQAFEAVGIDKAVIDEYFTGTPTQLGGIG
MSMEG_3225|M.smegmatis_MC2_155 CKGVVKVMSKMGISTVASYTAAQVFEAIGLDRGVIDEYFTGTTSQLGGVG
:**:*:********: **..** *:*:*:.. :::***:* . **:
Mb3889c|M.bovis_AF2122/97 LDDIAADVAARHRLAYLDRPDERAHRELEVGGEYQWRREGEYHLFNPETV
Rv3859c|M.tuberculosis_H37Rv LDDIAADVAARHRLAYLDRPDERAHRELEVGGEYQWRREGEYHLFNPETV
MLBr_00061|M.leprae_Br4923 LDDIAAEVAVRHRVAYLDRPDERAHRELEVGGEYQWRREGEYHLFNPETV
MMAR_5413|M.marinum_M LDDIAADVATRHTLAYLDRPDERAHRELEVGGEYQWRREGEYHLFNPETV
MUL_5037|M.ulcerans_Agy99 LDDIAADVASRHTLAYLDRPDERAHRELEVGGEYQWRREGEYHLFNPETV
MAV_0166|M.avium_104 LDDIAADVAARHALAYLDRPNERAHRELEVGGEYQWRREGEYHLFNPETV
TH_2102|M.thermoresistible__bu LDDIAADVARRHELAYLDRPEEWAHRELEVGGEYQWRREGEYHLFNPDTV
MAB_0091|M.abscessus_ATCC_1997 LDEIAADVASRHNLAFLDRPEEWAHRELEVGGEYQWRREGEYHLFNPDTV
Mflv_3593|M.gilvum_PYR-GCK LDVIAEEVKLRHRRAYPENPTERVHRRLEVGGEYAFRREGELHLFTPEVV
Mvan_2822|M.vanbaalenii_PYR-1 LDVIAEEVKLRHRRAYPENPTERVHRRLEVGGEYAFRREGELHLFTPEVV
MSMEG_3225|M.smegmatis_MC2_155 LDVLADEVKMRHRRAYPENPTERVHRRLEVGGEYAFRREGELHLFTPEVV
** :* :* ** *: :.* * .**.******* :***** ***.*:.*
Mb3889c|M.bovis_AF2122/97 FKLQHSTRTGQYKIFKEYTRLVDDQSERMASLRGLLKFRTGVRPPVPLDE
Rv3859c|M.tuberculosis_H37Rv FKLQHSTRTGQYKIFKEYTRLVDDQSERMASLRGLLKFRTGVRPPVPLDE
MLBr_00061|M.leprae_Br4923 FKLQHATRSGQYKIFKEYTRLVDDQSERMASLRGLLKFRSGVRTPVPLDE
MMAR_5413|M.marinum_M FKLQHSTRTGQYKIFKEYTRLVDDQSERMASLRGLLKFRAGVRPPIPLDE
MUL_5037|M.ulcerans_Agy99 FKLQHSTRTGQYKIFKEYTRLVDDQSERMASLRGLLKFRAGVRPPIPLDE
MAV_0166|M.avium_104 FKLQHATRTGQYKIFKDYTRLVDDQSERMASLRGLLKFRAGVRPPVPLEE
TH_2102|M.thermoresistible__bu FKLQHSTRTGQYEIFKEYTRLVDDQSERMASLRGLLKFKEGVRPPVPIEE
MAB_0091|M.abscessus_ATCC_1997 FKLQHSTRTGQYSVFKEYTQLVDDQSERMASLRGLLKFKTGVRPPVPLDE
Mflv_3593|M.gilvum_PYR-GCK FLLQHSTRTGRYEVFKQYSDEVDRLGRDGGALRGLFDFKKGLREPVPLDE
Mvan_2822|M.vanbaalenii_PYR-1 FLLQHSTRTGRYEVFEQYSEEVNRLAREGGALRGLFEFKTGLRPPVPLHE
MSMEG_3225|M.smegmatis_MC2_155 FLLQHSTRTGRRDIFAKYSEEVDRLSRKGGTLRGLFELKTGVRPPVPLEE
* ***:**:*: .:* .*: *: .. .:****:.:: *:* *:*:.*
Mb3889c|M.bovis_AF2122/97 VEPASEIVKRFSTGAMSYGSISAEAHETLAIAMNRLGARSNCGEGGEDVK
Rv3859c|M.tuberculosis_H37Rv VEPASEIVKRFSTGAMSYGSISAEAHETLAIAMNRLGARSNCGEGGEDVK
MLBr_00061|M.leprae_Br4923 VEPASEIVKRFSTGAMSYGSISAEAHETLAIAMNRLGGRSNSGEGGEDVN
MMAR_5413|M.marinum_M VEPASEIVKRFSTGAMSFGSISAEAHETLAIAMNRLGGRSNSGEGGEDVG
MUL_5037|M.ulcerans_Agy99 VEPASEIVKRFSTGAMSFGSISAEAHETLAIAMNRLGGRSNSGEGGEDVG
MAV_0166|M.avium_104 VEPASEIVKRFSTGAMSYGSISAEAHETLAIAMNRLGGRSNSGEGGEDVK
TH_2102|M.thermoresistible__bu VEPASEIVKRFSTGAMSYGSISAEAHETLAIAMNRLGARSNSGEGGEHVS
MAB_0091|M.abscessus_ATCC_1997 VEPASEIVKRFSTGAMSFGSISAEAHETLAIAMNRLGGRSNSGEGGEDPR
Mflv_3593|M.gilvum_PYR-GCK VESADEIVKRFNTGAMSYGSISAEAHETMAIAMNALGGRSNSGEGGEDVD
Mvan_2822|M.vanbaalenii_PYR-1 VESVDSIVTRFNTGAMSYGSISAEAHETMAVAMNNLGGRSNSGEGGEDVE
MSMEG_3225|M.smegmatis_MC2_155 VEPVESIVTRFNTGAMSYGSISAEAHETMAIAMNKLGGRSNSGEGGEDVD
**....**.**.*****:**********:*:*** **.***.*****.
Mb3889c|M.bovis_AF2122/97 RFDRDPNGDWRRSAIKQVASARFGVTSHYLTNCTDLQIKMAQGAKPGEGG
Rv3859c|M.tuberculosis_H37Rv RFDRDPNGDWRRSAIKQVASARFGVTSHYLTNCTDLQIKMAQGAKPGEGG
MLBr_00061|M.leprae_Br4923 RFEPDPNGDWRRSAIKQVASARFGVTSHYLTNCTDIQIKMAQGAKPGEGG
MMAR_5413|M.marinum_M RFEQDPNGDWRRSAIKQVASARFGVTSHYLTNCTDIQIKMAQGAKPGEGG
MUL_5037|M.ulcerans_Agy99 RFEQDPNGDWRRSAIKQVASARFGVTSHYLTNCTDIQIKMAQGAKPGEGG
MAV_0166|M.avium_104 RFDRDPDGSWRRSAIKQVASGRFGVTSHYLTNCTDIQIKMAQGAKPGEGG
TH_2102|M.thermoresistible__bu RFDPDDNGDWRRSAIKQVASGRFGVTSHYLTNCTDIQIKMAQGAKPGEGG
MAB_0091|M.abscessus_ATCC_1997 RFTPDENGDWRRSAIKQVASGRFGVTSHYLSNCTDIQIKMAQGAKPGEGG
Mflv_3593|M.gilvum_PYR-GCK RLYDPK----RRSAVKQVASGRFGVTSDYLVNATDIQIKMAQGAKPGEGG
Mvan_2822|M.vanbaalenii_PYR-1 RLYDPK----RRSAVKQVASGRFGVTSDYLVNATDIQIKMAQGAKPGEGG
MSMEG_3225|M.smegmatis_MC2_155 RLYDPR----RRSAVKQVASGRFGVTSDYLVNATDIQIKMAQGAKPGEGG
*: ****:*****.******.** *.**:**************
Mb3889c|M.bovis_AF2122/97 QLPGHKVYPWVAEVRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANP
Rv3859c|M.tuberculosis_H37Rv QLPGHKVYPWVAEVRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANP
MLBr_00061|M.leprae_Br4923 QLPGRKVYPWIAQVRNSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANP
MMAR_5413|M.marinum_M QLPGHKVYPWVAKVRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANP
MUL_5037|M.ulcerans_Agy99 QLPGHKVYPWVAKVRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANP
MAV_0166|M.avium_104 QLPGHKVYPWVAEVRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANP
TH_2102|M.thermoresistible__bu QLPGNKVYPWVAEVRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANP
MAB_0091|M.abscessus_ATCC_1997 QLPAHKVYPWVAEVRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNSNP
Mflv_3593|M.gilvum_PYR-GCK QLPGYKVYPNIAKTRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANA
Mvan_2822|M.vanbaalenii_PYR-1 QLPGYKVYPNIAKTRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANA
MSMEG_3225|M.smegmatis_MC2_155 QLPGYKVYPNIAKTRHSTPGVGLISPPPHHDIYSIEDLAQLIHDLKNANS
***. **** :*:.*:*******************************:*.
Mb3889c|M.bovis_AF2122/97 SARVHVKLVSENGVGTVAAGVSKAHADVVLISGHDGGTGATPLTSMKHAG
Rv3859c|M.tuberculosis_H37Rv SARVHVKLVSENGVGTVAAGVSKAHADVVLISGHDGGTGATPLTSMKHAG
MLBr_00061|M.leprae_Br4923 SARVHVKLVSENGVGTVAAGVSKAHADVVLVSGHDGGTGATPLTSMKHSG
MMAR_5413|M.marinum_M SARVHVKLVSENGVGTVAAGVSKAHADVVLISGHDGGTGATPLTSMKHAG
MUL_5037|M.ulcerans_Agy99 SARVHVKLVSENGVGTVAAGVSKAHADVVLISGHDGGTGATPLTSMKHAG
MAV_0166|M.avium_104 AARVHVKLVSENGVGTVAAGVSKAHADVVLISGHDGGTGATPLTSMKHAG
TH_2102|M.thermoresistible__bu QARVHVKLVSEAGVGTVAAGVSKAHADVVLISGHDGGTGASPLTSLKHAG
MAB_0091|M.abscessus_ATCC_1997 QARIHVKLVSENGVGTVATGVSKAHADVVLISGHDGGTGATPLTSMKHAG
Mflv_3593|M.gilvum_PYR-GCK DARIHVKLVSSVGVGTVAAGVSKAHADVVLISGYDGGTGAAPLTSLKHAG
Mvan_2822|M.vanbaalenii_PYR-1 DARIHVKLVSSVGVGTVAAGVSKAHADVVLISGYDGGTGAAPLTSLKHAG
MSMEG_3225|M.smegmatis_MC2_155 DARIHVKLVSSVGVGTVAAGVSKAHADVVLISGHDGGTGAAPLTSLKHAG
**:******. ******:***********:**:******:****:**:*
Mb3889c|M.bovis_AF2122/97 APWELGLAETQQTLLLNGLRDRIVVQVDGQLKTGRDVMIATLLGAEEFGF
Rv3859c|M.tuberculosis_H37Rv APWELGLAETQQTLLLNGLRDRIVVQVDGQLKTGRDVMIATLLGAEEFGF
MLBr_00061|M.leprae_Br4923 APWELGLAETQQTLLLNGLRDRIVVQVDGQLKTGRDVMIAALLGAEEFGF
MMAR_5413|M.marinum_M APWELGLAETQQTLLLNGLRDRIVVQVDGQLKTGRDVMIATLLGGEEFGF
MUL_5037|M.ulcerans_Agy99 APWELGLAETQQTLLLNGLRDRIVVQVDGQLKTGRDVMIATLLGGEEFGF
MAV_0166|M.avium_104 APWELGLAETQQTLLLNGLRDRIVVQVDGQLKTGRDVMIAALLGAEEFGF
TH_2102|M.thermoresistible__bu APWELGLAETQQTLLLNGLRDRIVVQVDGQLKTGRDVVIAALLGAEEFGF
MAB_0091|M.abscessus_ATCC_1997 APWELGLAETQQTLLLNGLRDRIVVQVDGQLKTGRDVMIAMLLGGEEFGF
Mflv_3593|M.gilvum_PYR-GCK APWEIGLADTQQTMVLNGLRDRITVQCDGGMRTARDVMVAMLLGAEEYGF
Mvan_2822|M.vanbaalenii_PYR-1 APWEIGLADTQQTLMLNGLRDRITVQCDGGMRTARDVMVAMLLGAEEYGF
MSMEG_3225|M.smegmatis_MC2_155 APWEIGLADTQQTLVLNGLRDRITVQCDGGLRTARDVVVAMLLGAEEYGF
****:***:****::********.** ** ::*.***::* ***.**:**
Mb3889c|M.bovis_AF2122/97 ATAPLVVAGCIMMRVCHLDTCPVGVATQNPLLRERFTGKPEFVENFFMFI
Rv3859c|M.tuberculosis_H37Rv ATAPLVVAGCIMMRVCHLDTCPVGVATQNPLLRERFTGKPEFVENFFMFI
MLBr_00061|M.leprae_Br4923 ATAPLVVAGCIMMRVCHLDTCPVGVATQNPLLRKRFNGKPEFVENFFMFI
MMAR_5413|M.marinum_M ATAPLVVAGCIMMRVCHLDTCPVGVATQNPLLRERFNGKPEFVENFFMFI
MUL_5037|M.ulcerans_Agy99 ATAPLVVAGCIMMRVCHLDTCPVGVATQNPLLRERFNGKPEFVENFFMFI
MAV_0166|M.avium_104 ATAPLVVSGCIMMRVCHLDTCPVGVATQNPVLRERFTGKPEFVENFFMFI
TH_2102|M.thermoresistible__bu ATAPLVVSGCVMMRVCHLDTCPVGVATQNPVLRQRFTGKPEFVENFFLFI
MAB_0091|M.abscessus_ATCC_1997 ATAPLVVSGCIMMRVCHLDTCPVGVATQNPVLRQRFNGKPEFVENFFLFI
Mflv_3593|M.gilvum_PYR-GCK ATAPLVVSGCIMMRVCHLDTCPVGVATQNPQLRARFNGKPEFVENFFRFI
Mvan_2822|M.vanbaalenii_PYR-1 ATAPLVVSGCIMMRVCHLDTCPVGVATQNPELRARFNGKPEFVENYFRFI
MSMEG_3225|M.smegmatis_MC2_155 ATAPLVVSGCIMMRVCHLDTCPVGVATQNPELRARFNGKPEFVENFFTFI
*******:**:******************* ** **.********:* **
Mb3889c|M.bovis_AF2122/97 AEEVREYLAQLGFRTVNEAVGQAGALDTTLARAHWKAHKLDLAPVLHEPE
Rv3859c|M.tuberculosis_H37Rv AEEVREYLAQLGFRTVNEAVGQAGALDTTLARAHWKAHKLDLAPVLHEPE
MLBr_00061|M.leprae_Br4923 AEEVREYMAQLGFRTFNEAVGQVGALDTTPVREHWKAHSIDLTPVLHEPE
MMAR_5413|M.marinum_M AEEVREYMAQLGFRTINEAVGQAGALDTTLARAHWKAHRLDLTPVLHEPE
MUL_5037|M.ulcerans_Agy99 AEEVREYMAQLGFRTINEAVGQAGALDTTLARAHWKAHRLDLTPVLHEPE
MAV_0166|M.avium_104 AEEVREYMAQLGFRTLNEAVGQVKSLDTTLARAHWKAHRLDLGPVLHEPE
TH_2102|M.thermoresistible__bu AEEVREYMAKMGFRTVNEMVGQVGMLDTTRAAEHWKAHKLDLSPVLHQPE
MAB_0091|M.abscessus_ATCC_1997 AEEVRELMAELGFRTVNEAVGQVGALDIERAVAHWKASKIDLTPVLTEPE
Mflv_3593|M.gilvum_PYR-GCK AEDIRKYLAELGFRSIDEAVGHAEMLDTAAGVAHWKSRGLDLSPLFAVPD
Mvan_2822|M.vanbaalenii_PYR-1 AEDIRKYLAELGFRSIDEAVGRAELLDTDMGVAHWKSKGLDLSPIFAVPS
MSMEG_3225|M.smegmatis_MC2_155 AEDIRRYLAELGFRSIDEAVGHAELLDTAEGVAHWKTRGLDLDPIFAVPT
**::*. :*::***:.:* **:. ** ***: :** *:: *
Mb3889c|M.bovis_AF2122/97 SAFMN--QDLYCSSRQDHGLDKALDQQLIVMSREALDSGKPVRFSTTIGN
Rv3859c|M.tuberculosis_H37Rv SAFMN--QDLYCSSRQDHGLDKALDQQLIVMSREALDSGKPVRFSTTIGN
MLBr_00061|M.leprae_Br4923 SAFMN--QDLYCSSRQDHGLDKVLDQQLIAVSRDALDFGKPVRFSTTIGN
MMAR_5413|M.marinum_M SAFMN--QDLYCSSSQDHGLDKALDQQLIVMSREALDSGKPVRFATTISN
MUL_5037|M.ulcerans_Agy99 SAFMN--QGLYCSSSQDHGLDKALDQQLIVMSREALDSGKPVRFATTISN
MAV_0166|M.avium_104 SAFMN--QDLYCSSRQDHGLDKALDQQLIVMSREALDSGKPVRFSTTISN
TH_2102|M.thermoresistible__bu SAFMN--QDLYCSSRQDHGLDKALDQQLITMCREALDNGTPVRFSTTIAN
MAB_0091|M.abscessus_ATCC_1997 SAFMN--QDLYCSGSQDHGLEKALDQQLIVMSREALDHGTPVKFETLITN
Mflv_3593|M.gilvum_PYR-GCK TASQ-----RRRTRDQDHGLDQALDRTLIQLAEGALEDAHPVRLELPVRN
Mvan_2822|M.vanbaalenii_PYR-1 NAHTGEPAARRRVRDQYHALDQALDQTLIQLAEGALEDAHPVRLELPIRN
MSMEG_3225|M.smegmatis_MC2_155 DAHGAKLTQRRKLREQDHGLDQALDRTLIQLAEGALEDAHPVRLELPVRN
* * *.*::.**: ** :.. **: . **:: : *
Mb3889c|M.bovis_AF2122/97 VNRTVGTMLGHELTKAYGGQGLPDGTIDITFDGSAGNSFGAFVPKGITLR
Rv3859c|M.tuberculosis_H37Rv VNRTVGTMLGHELTKAYGGQGLPDGTIDITFDGSAGNSFGAFVPKGITLR
MLBr_00061|M.leprae_Br4923 VNRTVGTMLGHELTKAYGAQGLPDGTIDITFEGSAGNSFGAFVPKGITLR
MMAR_5413|M.marinum_M VNRTVGTMLGHEVTKAYGGQGLPDGSIDITFEGSAGNSFGAFVPKGITLR
MUL_5037|M.ulcerans_Agy99 VNRTVGTMLGHEVTKAYGGQGLPDGSIDITFEGSAGNSFGAFVPKGITLR
MAV_0166|M.avium_104 VNRTVGTMLGHEVTKAYGGQGLPDGTIDITFDGSAGNSFGAFVPRGITLR
TH_2102|M.thermoresistible__bu VNRTVGTMLGHEVTKAYGGQGLPDGTIDITFDGSAGNSFGAFVPRGITLR
MAB_0091|M.abscessus_ATCC_1997 VNRTVGTMLGHEVTKAYGGEGLPDDTIDITFTGSAGNSFGAFVPRGITLR
Mflv_3593|M.gilvum_PYR-GCK VNRTVGTLLGSEVTRRYGAQGLPEDTIHIALTGSAGQSLGAFLPPGITLE
Mvan_2822|M.vanbaalenii_PYR-1 VNRTVGTLLGSEVTRRYGAQGLPDDSIHVTLTGSAGQSIGAFLPPGITLE
MSMEG_3225|M.smegmatis_MC2_155 VNRTVGTLLGSEVTRRYGAAGLPDDTIHVTLTGSAGQSIGAFLPPGITLE
*******:** *:*: **. ***:.:*.::: ****:*:***:* ****.
Mb3889c|M.bovis_AF2122/97 VYGDANDYVGKGLSGGRIVVRPSDDAPQDYVAEDNIIGGNVILFGATSGE
Rv3859c|M.tuberculosis_H37Rv VYGDANDYVGKGLSGGRIVVRPSDDAPQDYVAEDNIIGGNVILFGATSGE
MLBr_00061|M.leprae_Br4923 VYGDANDYVGKGLSGGRIVVRPSDSVPLNYVAEDNIIGGNVILFGATSGE
MMAR_5413|M.marinum_M IYGDANDYVGKGLSGGRIVVRPSDNAPPGYVAEDNIIGGNVILFGATSGE
MUL_5037|M.ulcerans_Agy99 IYGDANDYVGKGLSGGRIVVRPSDNAPPGYVAEDNIIGGNVILFGATSGE
MAV_0166|M.avium_104 VYGDANDYVGKGLSGGRIVVRPSDNAPADYVAEDNIIGGNVILFGATSGE
TH_2102|M.thermoresistible__bu VYGDANDYVGKGLSGGRIVVRPPDNVPESFVAEDNIIAGNVILFGATSGE
MAB_0091|M.abscessus_ATCC_1997 LFGDANDYVGKGLSGGHIVVRPSREAPEGFVAEKNIIGGNVILFGATSGE
Mflv_3593|M.gilvum_PYR-GCK LVGDANDYVGKGLSGGKIIVKPQDDV--LFLAEDNVIAGNTLLFGATSGE
Mvan_2822|M.vanbaalenii_PYR-1 LVGDANDYVGKGLSGGKIIVKPQDDV--LFLPEDNVIAGNTLLYGATSGE
MSMEG_3225|M.smegmatis_MC2_155 LVGDANDYVGKGLSGGRVVVRPPDDV--LFLPEDNVIAGNTLMYGATSGE
: **************:::*:* .. ::.*.*:*.**.:::******
Mb3889c|M.bovis_AF2122/97 VYLRGVVGERFAVRNSGAHAVVEGVGDHGCEYMTGGRVVILGRTGRNFAA
Rv3859c|M.tuberculosis_H37Rv VYLRGVVGERFAVRNSGAHAVVEGVGDHGCEYMTGGRVVILGRTGRNFAA
MLBr_00061|M.leprae_Br4923 VYLRGVVGERFAVRNSGAHAVVEGVGDHGCEYMTGGKVVILGRTGRNFAA
MMAR_5413|M.marinum_M VFLRGVVGERFAVRNSGAHAVVEGVGDHGCEYMTGGRVVILGQTGRNFAA
MUL_5037|M.ulcerans_Agy99 VFLRGVVGERFAVRNSGAHVVVEGVGDHGCEYMTGGRVVILGQTGRNFAA
MAV_0166|M.avium_104 AYLRGVVGERFAVRNSGAHAVVEGVGDHGCEYMTGGKVVVLGRTGRNFAA
TH_2102|M.thermoresistible__bu VFLRGQVGERFAVRNSGALAVVEGVGDHGCEYMTGGRVVILGPVGRNFAA
MAB_0091|M.abscessus_ATCC_1997 AFLNGVVGERFAVRNSGAAAVVEGVGDHGCEYMTGGTVVVLGPTGRNFAA
Mflv_3593|M.gilvum_PYR-GCK LYLRGRVGERFAARNSGALTVVEGVGDHACEYMTGGRVVVLGKVGRNMAA
Mvan_2822|M.vanbaalenii_PYR-1 LYLRGRVGERFAARNSGALAVVEGVGDHACEYMTGGRVVVLGKIGRNMAA
MSMEG_3225|M.smegmatis_MC2_155 VYLRGRVGERFCARNSGALAVVEGVGDHACEYMTGGRVVILGKTGRNMAA
:*.* *****..***** .********.******* **:** ***:**
Mb3889c|M.bovis_AF2122/97 GMSGGVAYVYDPDGELPANLNSEMVELETLDEDDADWLHGTIQVHVDATD
Rv3859c|M.tuberculosis_H37Rv GMSGGVAYVYDPDGELPANLNSEMVELETLDEDDADWLHGTIQVHVDATD
MLBr_00061|M.leprae_Br4923 GMSGGEAYVYDSDGELPANLNFEMVELEPLDFDDAAWLYATIQAHVDATD
MMAR_5413|M.marinum_M GMSGGIAYVYDPDGELPDNLNTEMVDVETLDDDDSEFVHGIVQAHVDATD
MUL_5037|M.ulcerans_Agy99 GMSGGIAYVYDPDGELPDNLNTEMVDVETLDDDDSEFVHGIVQAHVDATD
MAV_0166|M.avium_104 GMSGGVAYIYDPPGEFPQHLNAEMVELESLDDDDIEWLHGMIQAHVDATD
TH_2102|M.thermoresistible__bu GMSGGIAYVYDPDGVLPGRLNREMVELESLDDEDREWLRGFIAAHVDATD
MAB_0091|M.abscessus_ATCC_1997 GMSGGVAYVYDPDKRLMDNLNDEMVDLDALDPDDQQVLRSLIEKHVAATD
Mflv_3593|M.gilvum_PYR-GCK GMSGGIAFVLGLD---PRRVNTAMVELQRLEPEDLAWLHGVVGQHARYTG
Mvan_2822|M.vanbaalenii_PYR-1 GMSGGIAFVLGLD---PSRVNTDMVELQALEPEDLAWLRDVVAQHAHYTG
MSMEG_3225|M.smegmatis_MC2_155 GMSGGIAFVLGLD---PARVNTEMVQLQRLDPEDLEWLHHVVTDHVRYTG
***** *:: . .:* **::: *: :* : : *. *.
Mb3889c|M.bovis_AF2122/97 SAVGQRILSDWSGQQRHFVKVMPRDYKRVLQAIALAERDGVDVDKAIMAA
Rv3859c|M.tuberculosis_H37Rv SAVGQRILSDWSGQQRHFVKVMPRDYKRVLQAIALAERDGVDVDKAIMAA
MLBr_00061|M.leprae_Br4923 SAVGQRILAGWSEQQRYFVKVMPKDYKRVLQAIALAERNGVDVNKVIMEP
MMAR_5413|M.marinum_M SAVGQRVLADWPQQQRHFVKVMPRDYKRVLQAIAEAERDGVDIDKAIMAA
MUL_5037|M.ulcerans_Agy99 SAVGQRVLADWPQQQRHFVKVMPRDYKRVLQAIAEAERDGVDIDKAIMAA
MAV_0166|M.avium_104 SAVGQRILGDWAQQQRHFVKVMPRDYKRVLQAIAEAERDGGDVDKAIMAA
TH_2102|M.thermoresistible__bu SAIGQRILNDWDEHVTHFAKVMPRDYKRVLTAIAEAEASGADVDEAIMAA
MAB_0091|M.abscessus_ATCC_1997 SAVGQRILADWSGHSDSFVKVMPRDYRRVLEAIADAELTGGDVNEAIMAA
Mflv_3593|M.gilvum_PYR-GCK STVATSLLSDWPRRSAQFTKIMPRDYQRVLQATRMAKAEGRDVDSAIMEA
Mvan_2822|M.vanbaalenii_PYR-1 STVATSVLSDWPRRSAQFTKVMPRDFQRVLQATRMAKAEGRDVDTAIMEA
MSMEG_3225|M.smegmatis_MC2_155 STLGTSLLSDWPRRSAQFTKIMPTDYQRVLQATRMAKAEGRDVDSAIMEA
*::. :* .* : *.*:** *::*** * *: * *:: .** .
Mb3889c|M.bovis_AF2122/97 AHG
Rv3859c|M.tuberculosis_H37Rv AHG
MLBr_00061|M.leprae_Br4923 AHG
MMAR_5413|M.marinum_M SHG
MUL_5037|M.ulcerans_Agy99 SHG
MAV_0166|M.avium_104 AHG
TH_2102|M.thermoresistible__bu ANG
MAB_0091|M.abscessus_ATCC_1997 ARG
Mflv_3593|M.gilvum_PYR-GCK SRG
Mvan_2822|M.vanbaalenii_PYR-1 SRG
MSMEG_3225|M.smegmatis_MC2_155 SRG
:.*