For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTATTGSEIRSFADVPLEGETPAAAATPEARDAQVSAAASAHGYAPDQLTWTTPEGIDVKPVYIAADRDE AVEAGYPLDSFPGAPPFIRGPYPTMYVNQPWTIRQYAGFSTAAESNAFYRRNLAAGQKGLSVAFDLATHR GYDSDHPRVQGDVGMAGVAIDSILDMRQLFDGIDLSTVSVSMTMNGAVLPILALYVVAAEEQGVPPEKLA GTIQNDILKEFMVRNTYIYPPKESMRIISDIFAYTSAKMPKFNSISISGYHIQEAGATADLELAYTLADG VEYIKAGLDAGLDIDKFAPRLSFFWGIGMNFFMEVAKLRAGRLLWSELVSEFEPKSEKSLSLRTHSQTSG WSLTAQDVFNNVARTCIEAMAATQGHTQSLHTNALDEALALPTDFSARIARNTQLLLQQESGTTRPIDPW GGSYYVEWLTHQLAERARAHIKEVAEHGGMAQAISEGIPKLRIEEAAARTQARIDSGAQPVIGVNKYQVD EDTEIEVLKVENSRVRAEQIAKLQQLRAERDEAATQAALDELTRAAAASGPAGEDGLGNNLLALAIDAAR AKATVGEISDALEKVYGRHQAEIRTISGVYRDEVGKGSNIASATELVNKFAEADGRRPRILVAKMGQDGH DRGQKVIATAFADIGFDVDVGSLFSTPEEVARQAADNDVHVVGVSSLAAGHLTLVPALRDALAEVGRPDI MIVVGGVIPPGDFDELYAAGATAIFPPGTVIADAAIGLLHKLAERLGYELS
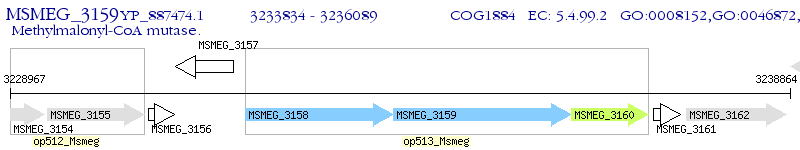
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3159 | - | - | 100% (751) | methylmalonyl-CoA mutase |
| M. smegmatis MC2 155 | MSMEG_4881 | - | e-110 | 43.45% (534) | methylmalonyl-CoA mutase, N-terminus of large subunit |
| M. smegmatis MC2 155 | MSMEG_3158 | mutA | 2e-29 | 29.27% (451) | methylmalonyl-CoA mutase, small subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1530 | mutB | 0.0 | 89.89% (742) | methylmalonyl-CoA mutase |
| M. gilvum PYR-GCK | Mflv_3649 | - | 0.0 | 91.66% (743) | methylmalonyl-CoA mutase |
| M. tuberculosis H37Rv | Rv1493 | mutB | 0.0 | 89.89% (742) | methylmalonyl-CoA mutase |
| M. leprae Br4923 | MLBr_01799 | mutB | 0.0 | 86.86% (746) | methylmalonyl-CoA mutase |
| M. abscessus ATCC 19977 | MAB_2711c | - | 0.0 | 87.07% (750) | methylmalonyl-CoA mutase |
| M. marinum M | MMAR_2303 | mutB | 0.0 | 90.40% (750) | methylmalonyl-CoA mutase large subunit, MutB |
| M. avium 104 | MAV_3277 | - | 0.0 | 89.69% (747) | methylmalonyl-CoA mutase |
| M. thermoresistible (build 8) | TH_0613 | mutB | 0.0 | 93.77% (690) | PROBABLE METHYLMALONYL-CoA MUTASE LARGE SUBUNIT MUTB (MCM) |
| M. ulcerans Agy99 | MUL_1506 | mutB | 0.0 | 89.87% (750) | methylmalonyl-CoA mutase |
| M. vanbaalenii PYR-1 | Mvan_2763 | - | 0.0 | 91.21% (751) | methylmalonyl-CoA mutase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3649|M.gilvum_PYR-GCK ----MTVTERRKS--------IDSFAEVPLLGDGAAAAPTQAEAAEQVTA
Mvan_2763|M.vanbaalenii_PYR-1 ----MTAIEPVK---------IDSFGDVPLHGDTAAAAPTYSDVTTQVSA
MSMEG_3159|M.smegmatis_MC2_155 ----MTATTGSE---------IRSFADVPLEGETPAAAATPEARDAQVSA
MAB_2711c|M.abscessus_ATCC_199 MTDLTDLSESGTI--------LGSFADVPLHGDRVSVAPDTAAVQTAVSE
TH_0613|M.thermoresistible__bu --------------------------------------------------
Mb1530|M.bovis_AF2122/97 ----MTTKTP----------VIGSFAGVPLHSERAAQSPTEAAVHTHVAA
Rv1493|M.tuberculosis_H37Rv ----MTTKTP----------VIGSFAGVPLHSERAAQSPTEAAVHTHVAA
MLBr_01799|M.leprae_Br4923 ----MTVRDVGGQQDRETTFVVGSFADIPLRSDRVVWPPSEAAVEKHVFS
MAV_3277|M.avium_104 ----MTTTAP----------AVGSFADVPLRSERPVAPATEAAVEEHVAA
MMAR_2303|M.marinum_M ----MTTTAS----------AIGSFADVPLHGERAVASATEAAVEKQVAA
MUL_1506|M.ulcerans_Agy99 ----MTTTAS----------AIGSFADVPLHGERAVASATEASVEKQVAA
Mflv_3649|M.gilvum_PYR-GCK AAAAHGYAADQSDWTTPEGIDVKPVFVAADRDDVVGAGYPLDSLPGEPPF
Mvan_2763|M.vanbaalenii_PYR-1 AAAAHGYTADQLDWATPEGIDVKPVYIGADRDAIVAAGYPLDSLPGEPPF
MSMEG_3159|M.smegmatis_MC2_155 AASAHGYAPDQLTWTTPEGIDVKPVYIAADRDEAVEAGYPLDSFPGAPPF
MAB_2711c|M.abscessus_ATCC_199 AAAAHGYSPEQLDWETPEAITVKPIYTAADREAVRAQGYPLDTFPGQVPF
TH_0613|M.thermoresistible__bu ------------------------VYIGADRAAAEAAGYPLHTFPGAVPY
Mb1530|M.bovis_AF2122/97 AAAAHGYTPEQLVWHTPEGIDVTPVYIAADRAAAEAEGYPLHSFPGEPPF
Rv1493|M.tuberculosis_H37Rv AAAAHGYTPEQLVWHTPEGIDVTPVYIAADRAAAEAEGYPLHSFPGEPPF
MLBr_01799|M.leprae_Br4923 AASAHGYTVDQLHWHTPEGIDVKPVYIAADRAAAEAGGYPLHSFPGEPPY
MAV_3277|M.avium_104 IAAAHLYTPDQLHWHTPEDIAVKPVYIGADRAAVEADGYPLHTFPGEPPF
MMAR_2303|M.marinum_M TAAAHGYATDQLRWQTPEGIEVKPVYIGADRRAAQADGYPVNSFPGEPPY
MUL_1506|M.ulcerans_Agy99 TAAAHGYATDQLRWQTPEGIEVKPVYIGADRRAAQADGYPVNSFPGEPPY
:: .*** ***:.::** *:
Mflv_3649|M.gilvum_PYR-GCK VRGPYPTMYVNQPWTIRQYAGFSTAAESNAFYRRNLAAGQKGLSVAFDLA
Mvan_2763|M.vanbaalenii_PYR-1 VRGPYPTMYVNQPWTIRQYAGFSTAAESNAFYRRNLAAGQKGLSVAFDLA
MSMEG_3159|M.smegmatis_MC2_155 IRGPYPTMYVNQPWTIRQYAGFSTAAESNAFYRRNLAAGQKGLSVAFDLA
MAB_2711c|M.abscessus_ATCC_199 LRGPYPTMYVNQPWTIRQYAGFSTAAESNAFYRRNLAAGQKGLSVAFDLA
TH_0613|M.thermoresistible__bu IRGPYPTMYVNQPWTIRQYAGFSTAAESNAFYRRNLAAGQKGLSVAFDLA
Mb1530|M.bovis_AF2122/97 VRGPYPTMYVNQPWTIRQYAGFSTAADSNAFYRRNLAAGQKGLSVAFDLA
Rv1493|M.tuberculosis_H37Rv VRGPYPTMYVNQPWTIRQYAGFSTAADSNAFYRRNLAAGQKGLSVAFDLA
MLBr_01799|M.leprae_Br4923 LRGPYPTMYVNQPWTIRQYAGFSTAADSNAFYRRNLAAGQKGLSVAFDLA
MAV_3277|M.avium_104 VRGPYPTMYVNQPWTIRQYAGFSTAAESNAFYRRNLAAGQKGLSVAFDLA
MMAR_2303|M.marinum_M VRGPYPTMYVNQPWTIRQYAGFSTAADSNAFYRRNLAAGQKGLSVAFDLA
MUL_1506|M.ulcerans_Agy99 VRGPYPTMYVNQPWTIRQYAGFSTAADSNAFYRRNLAAGQKGLSVAFDLA
:*************************:***********************
Mflv_3649|M.gilvum_PYR-GCK THRGYDSDHPRVAGDVGMAGVAIDSILDMRQLFDGIDLSSVSVSMTMNGA
Mvan_2763|M.vanbaalenii_PYR-1 THRGYDSDHPRVAGDVGMAGVAIDSILDMRQLFDGIDLSSVSVSMTMNGA
MSMEG_3159|M.smegmatis_MC2_155 THRGYDSDHPRVQGDVGMAGVAIDSILDMRQLFDGIDLSTVSVSMTMNGA
MAB_2711c|M.abscessus_ATCC_199 THRGYDSDHPRVQGDVGMAGVAIDSILDMRQLFDGIDLGSVSVSMTMNGA
TH_0613|M.thermoresistible__bu THRGYDSDHPRVQGDVGMAGVAIDSILDMRQLFDGIDLGSVSVSMTMNGA
Mb1530|M.bovis_AF2122/97 THRGYDSDHPRVQGDVGMAGVAIDSILDMRQLFDGIDLSTVSVSMTMNGA
Rv1493|M.tuberculosis_H37Rv THRGYDSDHPRVQGDVGMAGVAIDSILDMRQLFDGIDLSTVSVSMTMNGA
MLBr_01799|M.leprae_Br4923 THRGYDSDHPRVQGDVGMAGVAIDSILDMRQLFDGIDLSAVSVSMTMNGA
MAV_3277|M.avium_104 THRGYDSDHPRVQGDVGMAGVAIDSILDMRQLFDGIDLSTVSVSMTMNGA
MMAR_2303|M.marinum_M THRGYDSDHPRVQGDVGMAGVAIDSILDMRQLFDGIDLSSVSVSMTMNGA
MUL_1506|M.ulcerans_Agy99 THRGYDSDHPRVQGDVGMAGVAIDSILDMRQLFDGIDLSSVSVSMTMNGA
************ *************************.:**********
Mflv_3649|M.gilvum_PYR-GCK VLPILALYVVAAEEQGVPPEKLAGTIQNDILKEFMVRNTYIYPPKPSMRI
Mvan_2763|M.vanbaalenii_PYR-1 VLPILALYVVAAEEQGVPPEKLAGTIQNDILKEFMVRNTYIYPPKPSMRI
MSMEG_3159|M.smegmatis_MC2_155 VLPILALYVVAAEEQGVPPEKLAGTIQNDILKEFMVRNTYIYPPKESMRI
MAB_2711c|M.abscessus_ATCC_199 VLPILALYVVAAEEQGVGTEKLAGTIQNDILKEFMVRNTYIYPPKPSMRI
TH_0613|M.thermoresistible__bu VLPILALYVVAAEEQGVPPEKLAGTIQNDILKEFMVRNTYIYPPKPSMRI
Mb1530|M.bovis_AF2122/97 VLPILALYVVAAEEQGVAPEQLAGTIQNDILKEFMVRNTYIYPPKPSMRI
Rv1493|M.tuberculosis_H37Rv VLPILALYVVAAEEQGVAPEQLAGTIQNDILKEFMVRNTYIYPPKPSMRI
MLBr_01799|M.leprae_Br4923 VLPILALYVVAAEEQGVPPEKLAGTIQNDILKEFMVRNTYIYPPEPSMRI
MAV_3277|M.avium_104 VLPILALYVVAAEEQGVPPEKLAGTIQNDILKEFMVRNTYIYPPKPSMRI
MMAR_2303|M.marinum_M VLPILALYVVAAEEQGVPPEKLAGTIQNDILKEFMVRNTYIYPPKPSMRI
MUL_1506|M.ulcerans_Agy99 VLPILALYVVAAEEQGVPPEKLAGTIQNDILKEFMVRNTYIYPPKPSMRI
***************** .*:***********************: ****
Mflv_3649|M.gilvum_PYR-GCK ISDIFGYTSAKMPKFNSISISGYHIQEAGATADLELGYTLADGVEYIKAG
Mvan_2763|M.vanbaalenii_PYR-1 ISDIFGYTSAKMPKFNSISISGYHIQEAGATADLELGYTLADGVEYIKAG
MSMEG_3159|M.smegmatis_MC2_155 ISDIFAYTSAKMPKFNSISISGYHIQEAGATADLELAYTLADGVEYIKAG
MAB_2711c|M.abscessus_ATCC_199 ISDIFAYTSAKMPKFNSISISGYHIQEAGATADLELAYTLADGVEYIKAG
TH_0613|M.thermoresistible__bu ISDIFAYTSAKMPKFNSISISGYHIQEAGATADLELAYTLADGVEYIKAG
Mb1530|M.bovis_AF2122/97 ISDIFAYTSAKMPKFNSISISGYHIQEAGATADLELAYTLADGVDYIRAG
Rv1493|M.tuberculosis_H37Rv ISDIFAYTSAKMPKFNSISISGYHIQEAGATADLELAYTLADGVDYIRAG
MLBr_01799|M.leprae_Br4923 ISDIFAYTSAKMPKFNSISISGYHIQEAGATADLELAYTLADGVDYIKAG
MAV_3277|M.avium_104 ISDIFAYTSAKMPKFNSISISGYHIQEAGATADLELAYTLADGVDYIKAG
MMAR_2303|M.marinum_M ISDIFAYTSAKMPKFNSISISGYHIQEAGATADLELAYTLADGVDYIKAG
MUL_1506|M.ulcerans_Agy99 ISDIFAYTSVKMPKFNSISISGYHIQEAGATADLELAYTLADGVDYIKAG
*****.***.**************************.*******:**:**
Mflv_3649|M.gilvum_PYR-GCK LDAGLDIDKFAPRLSFFWGIGMNFFMEVAKLRAGRLLWSELVAEFDPKSS
Mvan_2763|M.vanbaalenii_PYR-1 LDAGLDIDKFAPRLSFFWGIGMNFFMEVAKLRAGRLLWSELVSQFEPKSA
MSMEG_3159|M.smegmatis_MC2_155 LDAGLDIDKFAPRLSFFWGIGMNFFMEVAKLRAGRLLWSELVSEFEPKSE
MAB_2711c|M.abscessus_ATCC_199 LAGGLDIDVFAPRLSFFWAIGMNFFMEVAKLRAGRLLWSELVAQFDPKSS
TH_0613|M.thermoresistible__bu LDAGLDIDKFAPRLSFFWGIGMNFFMEVAKLRAGRLLWSELVAQFDPKNS
Mb1530|M.bovis_AF2122/97 LNAGLDIDSFAPRLSFFWGIGMNFFMEVAKLRAGRLLWSELVAQFAPKSA
Rv1493|M.tuberculosis_H37Rv LNAGLDIDSFAPRLSFFWGIGMNFFMEVAKLRAGRLLWSELVAQFAPKSA
MLBr_01799|M.leprae_Br4923 LDAGLDIDTFAPRLSFFWGIGMNFFMEVAKLRAGRLLWSELVAKFGPQSP
MAV_3277|M.avium_104 LDAELDIDKFAPRLSFFWGIGMNFFMEVAKLRAGRLLWSELVSQFNPKNP
MMAR_2303|M.marinum_M LDAGLDIDKFAPRLSFFWGIGMNFFMEVAKLRAGRLLWSELVAKFEPKNS
MUL_1506|M.ulcerans_Agy99 LDAGLDIDKFAPRLSFFWGIGMNFFMEVAKLRAGRLLWSELVAKFEPKNS
* . **** *********.***********************::* *:.
Mflv_3649|M.gilvum_PYR-GCK KSLSLRTHSQTSGWSLTAQDAFNNVARTCIEAMAATQGHTQSLHTNALDE
Mvan_2763|M.vanbaalenii_PYR-1 KSLSLRTHSQTSGWSLTAQDAFNNVARTCVEAMAATQGHTQSLHTNALDE
MSMEG_3159|M.smegmatis_MC2_155 KSLSLRTHSQTSGWSLTAQDVFNNVARTCIEAMAATQGHTQSLHTNALDE
MAB_2711c|M.abscessus_ATCC_199 KSQSLRTHSQTSGWSLTAQDVFNNVARTCIEAMAATQGHTQSLHTNALDE
TH_0613|M.thermoresistible__bu KSLSLRTHSQTSGWSLTAQDVFNNVARTCIEAMAATQGHTQSLHTNALDE
Mb1530|M.bovis_AF2122/97 KSLSLRTHSQTSGWSLTAQDVFNNVARTCIEAMAATQGHTQSLHTNALDE
Rv1493|M.tuberculosis_H37Rv KSLSLRTHSQTSGWSLTAQDVFNNVARTCIEAMAATQGHTQSLHTNALDE
MLBr_01799|M.leprae_Br4923 KSLSLRTHSQTSGWSLTAQDPFNNVARTCIEAMAATQGHTQSLHTNALDE
MAV_3277|M.avium_104 KSLSLRTHSQTSGWSLTAQDAFNNVARTCIEAMAATQGHTQSLHTNALDE
MMAR_2303|M.marinum_M KSRSLRTHSQTSGWSLTAQDVFNNVARTCIEAMAATQGHTQSLHTNALDE
MUL_1506|M.ulcerans_Agy99 KSRSLRTHSQTSGWSLTAQDVFNNVARTCIEAMAATQGHTQSLHTNALDE
** ***************** ********:********************
Mflv_3649|M.gilvum_PYR-GCK ALALPTDFSARIARNTQLLLQQESGTTRPIDPWGGSYYVEWLTYQLAEKA
Mvan_2763|M.vanbaalenii_PYR-1 ALALPTDFSARIARNTQLLLQQESGTTRPIDPWAGSYYVESLTYQLAEKA
MSMEG_3159|M.smegmatis_MC2_155 ALALPTDFSARIARNTQLLLQQESGTTRPIDPWGGSYYVEWLTHQLAERA
MAB_2711c|M.abscessus_ATCC_199 ALALPTDFSARIARNTQLLIQQESGTCRPIDPWGGSYYVEWLTHQLASRA
TH_0613|M.thermoresistible__bu ALALPTDFSARIARNTQLLLQQESGTTRPIDPWGGSYYVEWLTHQLAERA
Mb1530|M.bovis_AF2122/97 ALALPTDFSARIARNTQLVLQQESGTTRPIDPWGGSYYVEWLTHRLARRA
Rv1493|M.tuberculosis_H37Rv ALALPTDFSARIARNTQLVLQQESGTTRPIDPWGGSYYVEWLTHRLARRA
MLBr_01799|M.leprae_Br4923 ALALPTDFSARIARNTQLVLQQESGTTRPIDPWGGSYYVEWLTHQLAARA
MAV_3277|M.avium_104 ALALPTDFSARIARNTQLVLQQESGTTRPIDPWGGSYYVEWLTHHLARKA
MMAR_2303|M.marinum_M ALALPTDFSARIARNTQLLLQQESGTTRPIDPWGGSYYVEWLTHQLAQRA
MUL_1506|M.ulcerans_Agy99 ALALPTDFSARIARNTQLLLQQESGTTRPIDPWAGSYYVEWLTHQLAQRA
******************::****** ******.****** **::** :*
Mflv_3649|M.gilvum_PYR-GCK RGHIREIAEHGGMAQAISDGIPKLRIEEAAARTQARIDSGQQPVIGVNKY
Mvan_2763|M.vanbaalenii_PYR-1 RAHIAEIAEYGGMAQAIDAGIPKLRIEEAAARTQARIDSGQQTVIGVNRY
MSMEG_3159|M.smegmatis_MC2_155 RAHIKEVAEHGGMAQAISEGIPKLRIEEAAARTQARIDSGAQPVIGVNKY
MAB_2711c|M.abscessus_ATCC_199 REHIREVTEAGGMAQAIDQGIPKLRIEEAAARTQARIDSGRQPVIGVNKY
TH_0613|M.thermoresistible__bu RAHIREVDEHGGMAQAISDGVPKLRIEEAAARTQARIDSGAQPVIGVNKY
Mb1530|M.bovis_AF2122/97 RAHIAEVAEHGGMAQAISDGIPKLRIEEAAARTQARIDSGQQPVVGVNKY
Rv1493|M.tuberculosis_H37Rv RAHIAEVAEHGGMAQAISDGIPKLRIEEAAARTQARIDSGQQPVVGVNKY
MLBr_01799|M.leprae_Br4923 RTHIAEVAEHGGMAAAISDGIPKLRIEEAAARTQARIDSGRQPVVGVNKY
MAV_3277|M.avium_104 RAHIAEVAEHGGMAQAISDGIPKLRIEEAAARTQARIDSGIQPVIGVNKY
MMAR_2303|M.marinum_M RAHINEVAELGGMAAAISEGIPKLRIEEAAARTQARIDSGQQPVIGVNKY
MUL_1506|M.ulcerans_Agy99 RAHINEVAELGGMAAAIGEGIPKLRIEEAAARTQARIDSGQQPVIGVNKY
* ** *: * **** **. *:******************* *.*:***:*
Mflv_3649|M.gilvum_PYR-GCK QVDEDHEIEVLKVENSRVRAEQLAKLERLRRERDETATQAALAELTRAAA
Mvan_2763|M.vanbaalenii_PYR-1 QVDEDHEIEVLKVENSRVRAEQIAKLERLRSERDDAATQAALAELTRAAA
MSMEG_3159|M.smegmatis_MC2_155 QVDEDTEIEVLKVENSRVRAEQIAKLQQLRAERDEAATQAALDELTRAAA
MAB_2711c|M.abscessus_ATCC_199 QVAEDQQIEVLKVENSRVRAEQLAKLQQLRAERDDDAVQAALAELTRAAA
TH_0613|M.thermoresistible__bu QVDEDHEIEVLKVDNSRVRAEQLAKLEQLRAERDQAAVDAALDELSRAAA
Mb1530|M.bovis_AF2122/97 QVPEDHEIEVLKVENSRVRAEQLAKLQRLRAGRDEPAVRAALAELTRAAA
Rv1493|M.tuberculosis_H37Rv QVPEDHEIEVLKVENSRVRAEQLAKLQRLRAGRDEPAVRAALAELTRAAA
MLBr_01799|M.leprae_Br4923 QVDQDHEIEVLKVENSRVRAEQVAKLQQLRANRDQLAVDAALAELSGAAG
MAV_3277|M.avium_104 QVEEDQEVEVLKVENSRVRAEQLAKLKELRDSRDQSAVDAALAELSRAAA
MMAR_2303|M.marinum_M QVDEDQQIEVLKVENSRVRAEQLAKLQELRAGRDQAAVDAALAELTRAAA
MUL_1506|M.ulcerans_Agy99 QVDEDQQIEVLKVENSRVRAEQLAKLQELRAGRDQAAVDAALAELTRAAA
** :* ::*****:********:***:.** **: *. *** **: **.
Mflv_3649|M.gilvum_PYR-GCK ASGVAGEDGLGNNLLALAIDAARAKATVGEISDALEKVYGRHQAEIRTIA
Mvan_2763|M.vanbaalenii_PYR-1 ASGAAGEDGLGNNLLALAIDAARAKATVGEISDALEKVYGRHQAEIRTIA
MSMEG_3159|M.smegmatis_MC2_155 ASGPAGEDGLGNNLLALAIDAARAKATVGEISDALEKVYGRHQAEIRTIS
MAB_2711c|M.abscessus_ATCC_199 DSGRAGEDGLGNNLMALAINAARHKATVGEISDALEKVYGRHQAEIRTIS
TH_0613|M.thermoresistible__bu ATGPAGADGLGNNLMALAINAARAKATVGEISSALERVWGRHQAEIRTIS
Mb1530|M.bovis_AF2122/97 EQGRAGADGLGNNLLALAIDAARAQATVGEISEALEKVYGRHRAEIRTIS
Rv1493|M.tuberculosis_H37Rv EQGRAGADGLGNNLLALAIDAARAQATVGEISEALEKVYGRHRAEIRTIS
MLBr_01799|M.leprae_Br4923 AHGRAG-----DNLLALAINAARVQATVGEISDALERVYGRHQAKIRTIT
MAV_3277|M.avium_104 AQGRAGEDGLGNNLLALAINAARAKATVGEISDALEKVYGRHQAEIRTIA
MMAR_2303|M.marinum_M AHGRAGDDGLGNNLLALAIDAARAKATVGEISDALEQVYGRHQAEIRTIA
MUL_1506|M.ulcerans_Agy99 AHGRAGDDGLGNNLLALAIDAARAKATVGEISDALEQVYGRHQAEIRTIA
* ** :**:****:*** :*******.***:*:***:*:****:
Mflv_3649|M.gilvum_PYR-GCK GVYRDEVG---MASNVSGATELVEKFAEADGRRPRILVAKMGQDGHDRGQ
Mvan_2763|M.vanbaalenii_PYR-1 GVYRDEVG---MAGNVSSATELVEKFAEADGRRPRILVAKMGQDGHDRGQ
MSMEG_3159|M.smegmatis_MC2_155 GVYRDEVG---KGSNIASATELVNKFAEADGRRPRILVAKMGQDGHDRGQ
MAB_2711c|M.abscessus_ATCC_199 GVYRDEVGRDGAVSSVTQATELVQKFADADGRRPRVLVAKMGQDGHDRGQ
TH_0613|M.thermoresistible__bu GVYRDEVG---KASNIASATELVEKFAEADGRRPRILVAKMGQDGHDRGQ
Mb1530|M.bovis_AF2122/97 GVYR---DEVGKAPNIAAATELVEKFAEADGRRPRILIAKMGQDGHDRGQ
Rv1493|M.tuberculosis_H37Rv GVYR---DEVGKAPNIAAATELVEKFAEADGRRPRILIAKMGQDGHDRGQ
MLBr_01799|M.leprae_Br4923 GVYRREVGEAGKATDIANATELVKKFAEADGRRPRILVAKMGQDGHDRGQ
MAV_3277|M.avium_104 GVYR---DEAGKATNIATATELVEKFAEADGRRPRILVAKMGQDGHDRGQ
MMAR_2303|M.marinum_M GVYRDEVQGGGNITPLSKATELVEKFAEADGRRPRILVAKMGQDGHDRGQ
MUL_1506|M.ulcerans_Agy99 GVYRDEVQGGGNITPLSKATELVEKFAEADGRRPRILVAKMGQDGHDRGQ
**** :: *****:***:*******:*:************
Mflv_3649|M.gilvum_PYR-GCK KVIATAFADIGFDVDVGSLFSTPDEVARQAADNDVHVVGVSSLAAGHLTL
Mvan_2763|M.vanbaalenii_PYR-1 KVIATAFADIGFDVDVGSLFSTPEEVARQAADNDVHVVGVSSLAAGHLTL
MSMEG_3159|M.smegmatis_MC2_155 KVIATAFADIGFDVDVGSLFSTPEEVARQAADNDVHVVGVSSLAAGHLTL
MAB_2711c|M.abscessus_ATCC_199 KVIATAFADIGFDVDVGPLFQTPEEVARQAADNDVHVVGVSSLAAGHLTL
TH_0613|M.thermoresistible__bu KVIATAFADIGFDVDVGSLFSTPDEVARQAADNDVHVVGVSSLAAGHLTL
Mb1530|M.bovis_AF2122/97 KVIATAFADIGFDVDVGSLFSTPEEVARQAADNDVHVIGVSSLAAGHLTL
Rv1493|M.tuberculosis_H37Rv KVIATAFADIGFDVDVGSLFSTPEEVARQAADNDVHVIGVSSLAAGHLTL
MLBr_01799|M.leprae_Br4923 KVVATAFADLGFDVDVGSLFSTPEEVARQAADNDVHVIGVSSLAAGHLTL
MAV_3277|M.avium_104 KVIATAFADIGFDVDVGSLFSTPEEVARQAADNDVHVVGVSSLAAGHLTL
MMAR_2303|M.marinum_M KVIATAFADIGFDVDVGSLFSTPEEVAREAADNDVHVVGVSSLAAGHLTL
MUL_1506|M.ulcerans_Agy99 KVIATAFADIGFDVDVGSLFSTPEEVAREAADNDVHVVGVSSLAAGHLTL
**:******:*******.**.**:****:********:************
Mflv_3649|M.gilvum_PYR-GCK VPALRDALAEVGRPDIMVVVGGVIPPGDFDELYEAGATAIFPPGTVIADA
Mvan_2763|M.vanbaalenii_PYR-1 VPALRDALAEVGRPDIMIVVGGVIPPGDFDELYAAGATAIFPPGTVIADA
MSMEG_3159|M.smegmatis_MC2_155 VPALRDALAEVGRPDIMIVVGGVIPPGDFDELYAAGATAIFPPGTVIADA
MAB_2711c|M.abscessus_ATCC_199 VPALRQALAEVGRPDIMIVVGGVIPPGDFDELYQAGAAAIFPPGTVISEA
TH_0613|M.thermoresistible__bu VPALRDALAEVGRPDIMIVVGGVIPPGDFEELYRAGATAIYPPGTVIADA
Mb1530|M.bovis_AF2122/97 VPALRDALAQVGRPDIMIVVGGVIPPGDFDELYAAGATAIFPPGTVIADA
Rv1493|M.tuberculosis_H37Rv VPALRDALAQVGRPDIMIVVGGVIPPGDFDELYAAGATAIFPPGTVIADA
MLBr_01799|M.leprae_Br4923 VPALRDALADVGRPDIMIVVGGVIPPDDFDELYAAGATAIFPPGTVIAYA
MAV_3277|M.avium_104 VPALRDALKEVGRPDIMIVVGGVIPPGDFDELYAAGAAAIFPPGTVIADA
MMAR_2303|M.marinum_M VPALRDALAKVGRPDIMIVVGGVIPPGDFDELYAAGATAIYPPGTVIADA
MUL_1506|M.ulcerans_Agy99 VPALRDALAKVGRPDIMIVVGGVIPPGDFDELYAAGATAIYPPGTVIADA
*****:** .*******:********.**:*** ***:**:******: *
Mflv_3649|M.gilvum_PYR-GCK AIGLLNKLAERLGYSLS
Mvan_2763|M.vanbaalenii_PYR-1 AIGLLNKLAERLGYTLS
MSMEG_3159|M.smegmatis_MC2_155 AIGLLHKLAERLGYELS
MAB_2711c|M.abscessus_ATCC_199 AIGLLHKLAERLGYDLT
TH_0613|M.thermoresistible__bu AIGLLHKLAERLGYDLS
Mb1530|M.bovis_AF2122/97 AIDLLHRLAERLGYTLD
Rv1493|M.tuberculosis_H37Rv AIDLLHRLAERLGYTLD
MLBr_01799|M.leprae_Br4923 AIGLLHKLAGRLGYTLT
MAV_3277|M.avium_104 AIGLLHKLAERLGYDLG
MMAR_2303|M.marinum_M AIGLLHKLAERLGYPLD
MUL_1506|M.ulcerans_Agy99 AIGLLHKLAERLGYPLD
**.**::** **** *