For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SEQAQPRVETPSGIPLAPVYGPQDRSGEPPPPGTYPFTRGNFASGYRGKTWTFRQYSGFGTAEESNRRYR YLLDQGGTGLSVALDLPTQCGYDSDDDEYGEEVGRVGVAVDTLADAEILFDGIPLEAISTSFTINGTAAI LLAFYVAAAEKKGVPREKLTGTIQNDILKEYASRGTWIWPPEPSLRLIADTIEFCAAEVPKFNAISVAGA HFRDAGANAVQEMAFTLADGVTYCDTVIERGRMTIDKFAPQISFFFYTHGDFFEEIAKYRAGRRRWATIV RERYGATTDKASMFRFGCVSGGASLYAPQAQNNLVRVAYEALASVLGGVQSMFTAAWDEPFALPSEESAT LALRTQQILAYETGVAKVADPLGGSYFVEALTDATEAKIVEIMSDLEAHGGMVACIEDGYLQGLIADEAY KLHQEIESGERPVVGVNRFVVDEPPPEIATYELDAEGRDLQLKRLSKVKAERDSAQVKATLAALSAAAEG DDNLMHKLIDCANAYCTVGEMVSALKAVWGEFQQPVVF*
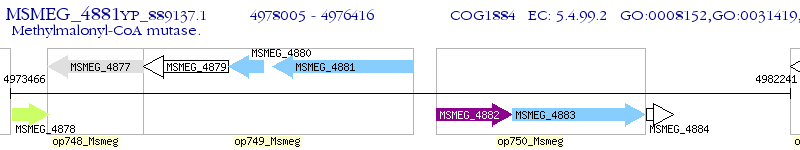
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4881 | - | - | 100% (529) | methylmalonyl-CoA mutase, N-terminus of large subunit |
| M. smegmatis MC2 155 | MSMEG_3159 | - | e-110 | 43.45% (534) | methylmalonyl-CoA mutase |
| M. smegmatis MC2 155 | MSMEG_3158 | mutA | 1e-23 | 27.19% (456) | methylmalonyl-CoA mutase, small subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1530 | mutB | 1e-108 | 42.32% (534) | methylmalonyl-CoA mutase |
| M. gilvum PYR-GCK | Mflv_2387 | - | 0.0 | 91.30% (529) | methylmalonyl-CoA mutase, large subunit |
| M. tuberculosis H37Rv | Rv1493 | mutB | 1e-108 | 42.32% (534) | methylmalonyl-CoA mutase |
| M. leprae Br4923 | MLBr_01799 | mutB | 1e-113 | 43.67% (529) | methylmalonyl-CoA mutase |
| M. abscessus ATCC 19977 | MAB_2711c | - | 1e-107 | 42.43% (535) | methylmalonyl-CoA mutase |
| M. marinum M | MMAR_4798 | mcmA2a | 0.0 | 86.35% (520) | methylmalonyl-CoA mutase beta subunit, McmA2a |
| M. avium 104 | MAV_0856 | - | 0.0 | 89.60% (519) | methylmalonyl-CoA mutase, N-terminus of large subunit |
| M. thermoresistible (build 8) | TH_0127 | - | 0.0 | 91.75% (521) | methylmalonyl-CoA mutase, N-terminus of large subunit |
| M. ulcerans Agy99 | MUL_0367 | mcmA2a | 0.0 | 85.96% (520) | methylmalonyl-CoA mutase beta subunit, McmA2a |
| M. vanbaalenii PYR-1 | Mvan_4261 | - | 0.0 | 92.25% (529) | methylmalonyl-CoA mutase, large subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2387|M.gilvum_PYR-GCK -----------------------MGTRTRCCVHLSLRRYWHSHFLQVRYP
Mvan_4261|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4881|M.smegmatis_MC2_155 --------------------------------------------------
TH_0127|M.thermoresistible__bu --------------------------------------------------
MAV_0856|M.avium_104 --------------------------------------------------
MMAR_4798|M.marinum_M --------------------------------------------------
MUL_0367|M.ulcerans_Agy99 --------------------------------------------------
Mb1530|M.bovis_AF2122/97 MTTKTP----------VIGSFAGVPLHSERAAQSPTEAAVHTHVAAAAAA
Rv1493|M.tuberculosis_H37Rv MTTKTP----------VIGSFAGVPLHSERAAQSPTEAAVHTHVAAAAAA
MLBr_01799|M.leprae_Br4923 MTVRDVGGQQDRETTFVVGSFADIPLRSDRVVWPPSEAAVEKHVFSAASA
MAB_2711c|M.abscessus_ATCC_199 MTDLTDLS----ESGTILGSFADVPLHGDRVSVAPDTAAVQTAVSEAAAA
Mflv_2387|M.gilvum_PYR-GCK AMSEQVVPRVETPSGIPLDPVYGPGDRAADPPP-------PGEYPFTRGN
Mvan_4261|M.vanbaalenii_PYR-1 -MSEQVHPLVETPSGIPLEPVYGPGDRAADPPP-------PGEYPFTRGN
MSMEG_4881|M.smegmatis_MC2_155 -MSEQAQPRVETPSGIPLAPVYGPQDRSGEPPP-------PGTYPFTRGN
TH_0127|M.thermoresistible__bu ---------VQTSSGIPLQPVYGPDDRPHEPPP-------PGTYPFTRGN
MAV_0856|M.avium_104 -----MDNTAHTPSGIPLQPVYGPADRSAEPPQ-------PGEFPFTRGN
MMAR_4798|M.marinum_M -----MDDVTQTASGIPVAPVYGPGDRTAEPPP-------PGEYPFTRGN
MUL_0367|M.ulcerans_Agy99 -----MDDVTQTASGIPVAPVYGPGDRTAEPPP-------PGEYPFTRGN
Mb1530|M.bovis_AF2122/97 HGYTPEQLVWHTPEGIDVTPVYIAADRAAAEAEGYPLHSFPGEPPFVRGP
Rv1493|M.tuberculosis_H37Rv HGYTPEQLVWHTPEGIDVTPVYIAADRAAAEAEGYPLHSFPGEPPFVRGP
MLBr_01799|M.leprae_Br4923 HGYTVDQLHWHTPEGIDVKPVYIAADRAAAEAGGYPLHSFPGEPPYLRGP
MAB_2711c|M.abscessus_ATCC_199 HGYSPEQLDWETPEAITVKPIYTAADREAVRAQGYPLDTFPGQVPFLRGP
.*...* : *:* . ** . ** *: **
Mflv_2387|M.gilvum_PYR-GCK FASGYRGKTWTFRQYSGFGTAEESNSRYRYLLDQGGTGLSVALDLPTQCG
Mvan_4261|M.vanbaalenii_PYR-1 FASGYRGKTWTFRQYSGFGTAEESNRRYRYLLEQGGTGLSVALDLPTQCG
MSMEG_4881|M.smegmatis_MC2_155 FASGYRGKTWTFRQYSGFGTAEESNRRYRYLLDQGGTGLSVALDLPTQCG
TH_0127|M.thermoresistible__bu FASGYRGKLWTFRQYSGFGTAEESNRRYRYLLEQGGTGLSVALDLPTQCG
MAV_0856|M.avium_104 FASGYRGKLWTFRQYSGFGTAEESNRRYRYLLEQGGTGLSVALDLPTQCG
MMAR_4798|M.marinum_M FASGYRGKLWTFRQYSGFGTAEESNRRYRYLLDQGGTGLSVALDLPTQCG
MUL_0367|M.ulcerans_Agy99 FASGYRGKLWTFRQYSGFGTAEESNRRYRYLLDQGGTGLSVALDLPTQCG
Mb1530|M.bovis_AF2122/97 YPTMYVNQPWTIRQYAGFSTAADSNAFYRRNLAAGQKGLSVAFDLATHRG
Rv1493|M.tuberculosis_H37Rv YPTMYVNQPWTIRQYAGFSTAADSNAFYRRNLAAGQKGLSVAFDLATHRG
MLBr_01799|M.leprae_Br4923 YPTMYVNQPWTIRQYAGFSTAADSNAFYRRNLAAGQKGLSVAFDLATHRG
MAB_2711c|M.abscessus_ATCC_199 YPTMYVNQPWTIRQYAGFSTAAESNAFYRRNLAAGQKGLSVAFDLATHRG
:.: * .: **:***:**.** :** ** * * .*****:**.*: *
Mflv_2387|M.gilvum_PYR-GCK YDSDDEEFGEEVGRVGVAVDTLADAEILFDGIPLDKISTSFTINGTAAIL
Mvan_4261|M.vanbaalenii_PYR-1 YDSDDAEYGEEVGRVGVAVDTLADAEILFDGIPLDKISTSFTINGTAAIL
MSMEG_4881|M.smegmatis_MC2_155 YDSDDDEYGEEVGRVGVAVDTLADAEILFDGIPLEAISTSFTINGTAAIL
TH_0127|M.thermoresistible__bu YDSDDPEYGEEVGRVGVAVDTLADAEILFDGIPLDKISTSFTINGTAAIL
MAV_0856|M.avium_104 YDSDDPEFGEEVGRVGVAVDTLADFEILFDGIPLDKLSTSMTINGTAAIL
MMAR_4798|M.marinum_M FDSDDPDVAEEVGRVGVAVDTLADAEILFQDIPLDKISTSFTINGTAAIL
MUL_0367|M.ulcerans_Agy99 FDSDDPDVAEEVGRVGVAVDTLADAEILLEDIPLDKISTSFTINGTAAIL
Mb1530|M.bovis_AF2122/97 YDSDHPRVQGDVGMAGVAIDSILDMRQLFDGIDLSTVSVSMTMNGAVLPI
Rv1493|M.tuberculosis_H37Rv YDSDHPRVQGDVGMAGVAIDSILDMRQLFDGIDLSTVSVSMTMNGAVLPI
MLBr_01799|M.leprae_Br4923 YDSDHPRVQGDVGMAGVAIDSILDMRQLFDGIDLSAVSVSMTMNGAVLPI
MAB_2711c|M.abscessus_ATCC_199 YDSDHPRVQGDVGMAGVAIDSILDMRQLFDGIDLGSVSVSMTMNGAVLPI
:***. :** .***:*:: * . *::.* * :*.*:*:**:. :
Mflv_2387|M.gilvum_PYR-GCK LAFYVAAAEKKGVPREKLTGTIQNDILKEYASRGTWIWPPEPSLRLIADT
Mvan_4261|M.vanbaalenii_PYR-1 LAFYVAAAEKKGVPREKLTGTIQNDILKEYASRGTWIWPPEPSLRLIADT
MSMEG_4881|M.smegmatis_MC2_155 LAFYVAAAEKKGVPREKLTGTIQNDILKEYASRGTWIWPPEPSLRLIADT
TH_0127|M.thermoresistible__bu LAFYVAAAEKTGVPRAKLTGTIQNDILKEYASRGTWIWPPEPSLRLIADT
MAV_0856|M.avium_104 LAFYVAAAEKKGIPRANLTGTIQNDILKEYASRGTWIWPPEPSLRLIADT
MMAR_4798|M.marinum_M LAFYVAAAERKGVPRAKLTGTIQNDILKEYASRGTWIWPPGPSLRLIADT
MUL_0367|M.ulcerans_Agy99 LAFYAAAAERKGVPRAKLTGTIQNDILKEYASRGTWIWPPEPSLRLIADT
Mb1530|M.bovis_AF2122/97 LALYVVAAEEQGVAPEQLAGTIQNDILKEFMVRNTYIYPPKPSMRIISDI
Rv1493|M.tuberculosis_H37Rv LALYVVAAEEQGVAPEQLAGTIQNDILKEFMVRNTYIYPPKPSMRIISDI
MLBr_01799|M.leprae_Br4923 LALYVVAAEEQGVPPEKLAGTIQNDILKEFMVRNTYIYPPEPSMRIISDI
MAB_2711c|M.abscessus_ATCC_199 LALYVVAAEEQGVGTEKLAGTIQNDILKEFMVRNTYIYPPKPSMRIISDI
**:*..***. *: :*:**********: *.*:*:** **:*:*:*
Mflv_2387|M.gilvum_PYR-GCK IEFCAAEVPRFNAISVAGAHFRDAGANAVQEMAFTLADGVTYCDTVVERG
Mvan_4261|M.vanbaalenii_PYR-1 IEFCAAEVPKFNAISVAGAHFRDAGANAVQEMAFTLADGVTYCDTVVERG
MSMEG_4881|M.smegmatis_MC2_155 IEFCAAEVPKFNAISVAGAHFRDAGANAVQEMAFTLADGVTYCDTVIERG
TH_0127|M.thermoresistible__bu IEFCAEEVPKFNAISVAGAHFRDAGANAVQEMAFTLADGVTYCDTVIERG
MAV_0856|M.avium_104 IEFCAAEVPKFNAISVAGAHFRDAGANAVQEMAFTLADGVTYCDTVVERG
MMAR_4798|M.marinum_M IEFCAAEVPKFNAISVAGAHFRDAGATAVQEMAFTLADGITYCDTVVARG
MUL_0367|M.ulcerans_Agy99 IEFCAAEVPKFNAISVAGAHFRDAGATAVQEMAFTLADGITHCDTVVVRG
Mb1530|M.bovis_AF2122/97 FAYTSAKMPKFNSISISGYHIQEAGATADLELAYTLADGVDYIRAGLNAG
Rv1493|M.tuberculosis_H37Rv FAYTSAKMPKFNSISISGYHIQEAGATADLELAYTLADGVDYIRAGLNAG
MLBr_01799|M.leprae_Br4923 FAYTSAKMPKFNSISISGYHIQEAGATADLELAYTLADGVDYIKAGLDAG
MAB_2711c|M.abscessus_ATCC_199 FAYTSAKMPKFNSISISGYHIQEAGATADLELAYTLADGVEYIKAGLAGG
: : : ::*:**:**::* *:::***.* *:*:*****: : : : *
Mflv_2387|M.gilvum_PYR-GCK RMTIDKFAPQISFFFYTHGDFFEEIAKYRAGRRRWATIVRERYGATTDKA
Mvan_4261|M.vanbaalenii_PYR-1 RMTIDKFAPQISFFFYTHGDFFEEIAKYRAGRRRWATIVRERYGATTDKA
MSMEG_4881|M.smegmatis_MC2_155 RMTIDKFAPQISFFFYTHGDFFEEIAKYRAGRRRWATIVRERYGATTDKA
TH_0127|M.thermoresistible__bu RMTIDQFAPQISFFFYTHGDFFEEIAKYRAGRRRWATIVRERYGAETDKA
MAV_0856|M.avium_104 RMTIDQFAPQISFFFYTHGDFFEEIAKYRAGRRRWATIVRERYGATTAKA
MMAR_4798|M.marinum_M RMSIDQFAPQISFFFYTHGDFFEEIAKYRAGRRRWATLVRERYGAKTDKA
MUL_0367|M.ulcerans_Agy99 RMTIDQFAPQISFFFYTHGDFFEEIAKYRAGRRRWATLVRERYGAKTDKA
Mb1530|M.bovis_AF2122/97 -LDIDSFAPRLSFFWGIGMNFFMEVAKLRAGRLLWSELVAQ-FAPKSAKS
Rv1493|M.tuberculosis_H37Rv -LDIDSFAPRLSFFWGIGMNFFMEVAKLRAGRLLWSELVAQ-FAPKSAKS
MLBr_01799|M.leprae_Br4923 -LDIDTFAPRLSFFWGIGMNFFMEVAKLRAGRLLWSELVAK-FGPQSPKS
MAB_2711c|M.abscessus_ATCC_199 -LDIDVFAPRLSFFWAIGMNFFMEVAKLRAGRLLWSELVAQ-FDPKSSKS
: ** ***::***: :** *:** **** *: :* : : . : *:
Mflv_2387|M.gilvum_PYR-GCK SMFRFGCVAGGASLYAPQAQNNLVRVAYEAMAAVLGGVQSMFTAAWDEPF
Mvan_4261|M.vanbaalenii_PYR-1 SMFRFGCVAGGASLYAPQAQNNLVRVAYEAMAAVLGGVQSMFTAAWDEPF
MSMEG_4881|M.smegmatis_MC2_155 SMFRFGCVSGGASLYAPQAQNNLVRVAYEALASVLGGVQSMFTAAWDEPF
TH_0127|M.thermoresistible__bu AMFRFGCVSGGASLYAPQAQNNLIRVAYEALASVLGGVQSMFTAAWDEPF
MAV_0856|M.avium_104 SMFRFGCVCGGASLYAPQAHNNIVRVAYEAMAAVLGGVQSMFTAAWDEPF
MMAR_4798|M.marinum_M AMFRFGCVCGGASLYAPQAQNNLIRVAYEAMAAVLGGVQSMFTAAWDEPF
MUL_0367|M.ulcerans_Agy99 AMFRFGCVCGGASLYAPQAQNNLIRAAYEAMAAVLGGVQPMFTAAWDEPF
Mb1530|M.bovis_AF2122/97 LSLRTHSQTSGWSLTAQDVFNNVARTCIEAMAATQGHTQSLHTNALDEAL
Rv1493|M.tuberculosis_H37Rv LSLRTHSQTSGWSLTAQDVFNNVARTCIEAMAATQGHTQSLHTNALDEAL
MLBr_01799|M.leprae_Br4923 LSLRTHSQTSGWSLTAQDPFNNVARTCIEAMAATQGHTQSLHTNALDEAL
MAB_2711c|M.abscessus_ATCC_199 QSLRTHSQTSGWSLTAQDVFNNVARTCIEAMAATQGHTQSLHTNALDEAL
:* . .* ** * : **: *.. **:*:. * .*.:.* * **.:
Mflv_2387|M.gilvum_PYR-GCK ALPSEESATLALRTQQILAYETGVTKVADPLGGSYFVEALTDATEEKIIE
Mvan_4261|M.vanbaalenii_PYR-1 ALPSEESATLALRTQQILAYETGVTKVADPLGGSYFVEALTDATEEKIIE
MSMEG_4881|M.smegmatis_MC2_155 ALPSEESATLALRTQQILAYETGVAKVADPLGGSYFVEALTDATEAKIVE
TH_0127|M.thermoresistible__bu ALPSEESATMALRTQQILAYETGVAKVADPLGGSYFVEALTDATEEKIIE
MAV_0856|M.avium_104 ALPTEETTTLALRTQQILAYETGVASVADPLGGSYFVEALTDETEARIIE
MMAR_4798|M.marinum_M ALPTEESTTLALRTQQILAYESGVTRVADPLGGSYFVEALTDATEARIIE
MUL_0367|M.ulcerans_Agy99 ALPTEESTTLALRTQQILAYESGVTRVADPLGGSYFVEALTDATEARIIE
Mb1530|M.bovis_AF2122/97 ALPTDFSARIARNTQLVLQQESGTTRPIDPWGGSYYVEWLTHRLARRARA
Rv1493|M.tuberculosis_H37Rv ALPTDFSARIARNTQLVLQQESGTTRPIDPWGGSYYVEWLTHRLARRARA
MLBr_01799|M.leprae_Br4923 ALPTDFSARIARNTQLVLQQESGTTRPIDPWGGSYYVEWLTHQLAARART
MAB_2711c|M.abscessus_ATCC_199 ALPTDFSARIARNTQLLIQQESGTCRPIDPWGGSYYVEWLTHQLASRARE
***:: :: :* .** :: *:*. ** ****:** **. :
Mflv_2387|M.gilvum_PYR-GCK IMHDLETHGGMVRCIEDGYLQGLIADEAFKIHREVESGERPVVGVNKFVV
Mvan_4261|M.vanbaalenii_PYR-1 IMADLEAHGGMVRCIEDGYLQGLIADEAFKIHQEVESGVRPVVGVNKFVV
MSMEG_4881|M.smegmatis_MC2_155 IMSDLEAHGGMVACIEDGYLQGLIADEAYKLHQEIESGERPVVGVNRFVV
TH_0127|M.thermoresistible__bu IMADLEAHGGMVRAIEDGYLQGLIADEAYRVHREIESGERPVVGVNRFVS
MAV_0856|M.avium_104 IMADLDRHGGMVSAIEDGYLQGLIADEAFRLHQDIEAGTRPVVGVNRFVT
MMAR_4798|M.marinum_M IMDDLQRHGGMVSAIEDGYLQGLIADEAFRTHKDIEAGVRPVVGVNRFVS
MUL_0367|M.ulcerans_Agy99 IMDDLRRHGGMVSAIEDGYLQGLIADEAFRTHKDIEAGVRPVVGVNRFVS
Mb1530|M.bovis_AF2122/97 HIAEVAEHGGMAQAISDGIPKLRIEEAAARTQARIDSGQQPVVGVNKYQV
Rv1493|M.tuberculosis_H37Rv HIAEVAEHGGMAQAISDGIPKLRIEEAAARTQARIDSGQQPVVGVNKYQV
MLBr_01799|M.leprae_Br4923 HIAEVAEHGGMAAAISDGIPKLRIEEAAARTQARIDSGRQPVVGVNKYQV
MAB_2711c|M.abscessus_ATCC_199 HIREVTEAGGMAQAIDQGIPKLRIEEAAARTQARIDSGRQPVIGVNKYQV
: :: ***. .*.:* : * : * : : :::* :**:***::
Mflv_2387|M.gilvum_PYR-GCK DEPAPDLATYELDAEGRDKQLTRLAKVKAERDGVAVKESLAALARAAEGE
Mvan_4261|M.vanbaalenii_PYR-1 DEPPPDLATYELDAEGRDKQLKRLAKVKADRDDVAVKEALAALARAAEGD
MSMEG_4881|M.smegmatis_MC2_155 DEPPPEIATYELDAEGRDLQLKRLSKVKAERDSAQVKATLAALSAAAEGD
TH_0127|M.thermoresistible__bu DEPPPQISTYELDAEGRDLQLKRLTKVKAERDAGAVQAALAELSRAAEGT
MAV_0856|M.avium_104 EEPEHDVVTYELDAEGRDLQLKRLSKVKAERDSAAVKSSLAALSRAAEGD
MMAR_4798|M.marinum_M GDAEPDIVTYELDAEGRDLQLKRLAKVKAERDSDAVQSSLAALARGAEGA
MUL_0367|M.ulcerans_Agy99 GDAEPDIVTYELDAEGRDLQLKRLAKVKAERDSDAVQSTLAALARGAEGP
Mb1530|M.bovis_AF2122/97 PEDHEIEVLKVENSRVRAEQLAKLQRLRAGRDEPAVRAALAELTRAAAEQ
Rv1493|M.tuberculosis_H37Rv PEDHEIEVLKVENSRVRAEQLAKLQRLRAGRDEPAVRAALAELTRAAAEQ
MLBr_01799|M.leprae_Br4923 DQDHEIEVLKVENSRVRAEQVAKLQQLRANRDQLAVDAALAELSGAAGAH
MAB_2711c|M.abscessus_ATCC_199 AEDQQIEVLKVENSRVRAEQLAKLQQLRAERDDDAVQAALAELTRAAADS
: ::. * *: :* :::* ** * :** *: .*
Mflv_2387|M.gilvum_PYR-GCK ---------DNLMHKLIDCANVYCTVGEMVSTLKSVWGEFQQPVVF----
Mvan_4261|M.vanbaalenii_PYR-1 ---------DNLMHKLIDCANAYCTVGEMVSTLKSVWGEFQQPVVF----
MSMEG_4881|M.smegmatis_MC2_155 ---------DNLMHKLIDCANAYCTVGEMVSALKAVWGEFQQPVVF----
TH_0127|M.thermoresistible__bu ---------ENLMPKLIDCANAYCTVGEMVTALKKVWGEFQQPVVF----
MAV_0856|M.avium_104 ---------DNLMHKLIDCANAYCTVGEMVSALKAVWGEFQQPVVF----
MMAR_4798|M.marinum_M ---------DNLMHRLIDCANAYCTVGEMVCALKAVWGEFQQPVVF----
MUL_0367|M.ulcerans_Agy99 ---------DNLMHRLIDCANAYCTVGEMVCALKAVWGEFQQPVVF----
Mb1530|M.bovis_AF2122/97 GRAGADGLGNNLLALAIDAARAQATVGEISEALEKVYGRHRAEIRTISGV
Rv1493|M.tuberculosis_H37Rv GRAGADGLGNNLLALAIDAARAQATVGEISEALEKVYGRHRAEIRTISGV
MLBr_01799|M.leprae_Br4923 GRAG-----DNLLALAINAARVQATVGEISDALERVYGRHQAKIRTITGV
MAB_2711c|M.abscessus_ATCC_199 GRAGEDGLGNNLMALAINAARHKATVGEISDALEKVYGRHQAEIRTISGV
:**: *:.*. .****: :*: *:*..: :
Mflv_2387|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4261|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4881|M.smegmatis_MC2_155 --------------------------------------------------
TH_0127|M.thermoresistible__bu --------------------------------------------------
MAV_0856|M.avium_104 --------------------------------------------------
MMAR_4798|M.marinum_M --------------------------------------------------
MUL_0367|M.ulcerans_Agy99 --------------------------------------------------
Mb1530|M.bovis_AF2122/97 YR---DEVGKAPNIAAATELVEKFAEADGRRPRILIAKMGQDGHDRGQKV
Rv1493|M.tuberculosis_H37Rv YR---DEVGKAPNIAAATELVEKFAEADGRRPRILIAKMGQDGHDRGQKV
MLBr_01799|M.leprae_Br4923 YRREVGEAGKATDIANATELVKKFAEADGRRPRILVAKMGQDGHDRGQKV
MAB_2711c|M.abscessus_ATCC_199 YRDEVGRDGAVSSVTQATELVQKFADADGRRPRVLVAKMGQDGHDRGQKV
Mflv_2387|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4261|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4881|M.smegmatis_MC2_155 --------------------------------------------------
TH_0127|M.thermoresistible__bu --------------------------------------------------
MAV_0856|M.avium_104 --------------------------------------------------
MMAR_4798|M.marinum_M --------------------------------------------------
MUL_0367|M.ulcerans_Agy99 --------------------------------------------------
Mb1530|M.bovis_AF2122/97 IATAFADIGFDVDVGSLFSTPEEVARQAADNDVHVIGVSSLAAGHLTLVP
Rv1493|M.tuberculosis_H37Rv IATAFADIGFDVDVGSLFSTPEEVARQAADNDVHVIGVSSLAAGHLTLVP
MLBr_01799|M.leprae_Br4923 VATAFADLGFDVDVGSLFSTPEEVARQAADNDVHVIGVSSLAAGHLTLVP
MAB_2711c|M.abscessus_ATCC_199 IATAFADIGFDVDVGPLFQTPEEVARQAADNDVHVVGVSSLAAGHLTLVP
Mflv_2387|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4261|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4881|M.smegmatis_MC2_155 --------------------------------------------------
TH_0127|M.thermoresistible__bu --------------------------------------------------
MAV_0856|M.avium_104 --------------------------------------------------
MMAR_4798|M.marinum_M --------------------------------------------------
MUL_0367|M.ulcerans_Agy99 --------------------------------------------------
Mb1530|M.bovis_AF2122/97 ALRDALAQVGRPDIMIVVGGVIPPGDFDELYAAGATAIFPPGTVIADAAI
Rv1493|M.tuberculosis_H37Rv ALRDALAQVGRPDIMIVVGGVIPPGDFDELYAAGATAIFPPGTVIADAAI
MLBr_01799|M.leprae_Br4923 ALRDALADVGRPDIMIVVGGVIPPDDFDELYAAGATAIFPPGTVIAYAAI
MAB_2711c|M.abscessus_ATCC_199 ALRQALAEVGRPDIMIVVGGVIPPGDFDELYQAGAAAIFPPGTVISEAAI
Mflv_2387|M.gilvum_PYR-GCK ---------------
Mvan_4261|M.vanbaalenii_PYR-1 ---------------
MSMEG_4881|M.smegmatis_MC2_155 ---------------
TH_0127|M.thermoresistible__bu ---------------
MAV_0856|M.avium_104 ---------------
MMAR_4798|M.marinum_M ---------------
MUL_0367|M.ulcerans_Agy99 ---------------
Mb1530|M.bovis_AF2122/97 DLLHRLAERLGYTLD
Rv1493|M.tuberculosis_H37Rv DLLHRLAERLGYTLD
MLBr_01799|M.leprae_Br4923 GLLHKLAGRLGYTLT
MAB_2711c|M.abscessus_ATCC_199 GLLHKLAERLGYDLT