For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VEPSDEFRHNGVVKLRGGATSPVATGPSVPAADEVPGVISTADGQTRRAIIKLLLEGPITAGQIGERLGI SAAGVRRHLDALIEAGDAQASAAAAWQHTGRGRPAKRYRLTAAGRAKLGHSYDDLAVAAIRQLREIGGKD AVRTFARRRIDTILSGVTAGSGDVAQTAERVADAMTKAGYATTTAPVSGPIRGVQICQHHCPVSHVAEEF PELCETEREAFAEILGTHVQRLATIVNGDCACTTHVPLDLDPPGEHTDHPKDVRSTARDRTTTPSDQGAS T
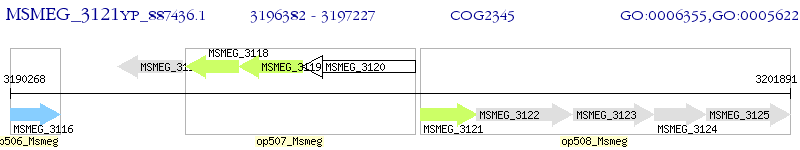
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3121 | - | - | 100% (281) | DNA-binding protein |
| M. smegmatis MC2 155 | MSMEG_1125 | - | 7e-05 | 27.49% (171) | regulatory protein, ArsR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1495 | - | 3e-93 | 69.26% (257) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_3678 | - | 6e-92 | 68.83% (247) | putative transcriptional regulator |
| M. tuberculosis H37Rv | Rv1460 | - | 3e-93 | 69.26% (257) | transcriptional regulatory protein |
| M. leprae Br4923 | MLBr_00592 | - | 4e-89 | 64.94% (251) | putative DNA-binding protein |
| M. abscessus ATCC 19977 | MAB_2750c | - | 1e-87 | 66.26% (243) | transcriptional regulatory protein |
| M. marinum M | MMAR_2265 | - | 3e-92 | 68.09% (257) | transcriptional regulatory protein |
| M. avium 104 | MAV_3319 | - | 5e-89 | 74.41% (211) | DNA-binding protein |
| M. thermoresistible (build 8) | TH_1094 | - | 1e-45 | 67.86% (140) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN |
| M. ulcerans Agy99 | MUL_1861 | - | 2e-92 | 68.09% (257) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_2732 | - | 8e-88 | 76.19% (210) | putative transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1495|M.bovis_AF2122/97 MTSTTLPHRASLVDRSTEFCHTDVVKIP-------------AVSTTV--P
Rv1460|M.tuberculosis_H37Rv MTSTTLPHRASLVDRSTEFCHTDVVKIP-------------AVSTTV--P
MAV_3319|M.avium_104 --------------------------MG-------------AAG------
MMAR_2265|M.marinum_M -----------MLDRCPELRHTGVVKIQPEFS---------AAHSAA--L
MUL_1861|M.ulcerans_Agy99 -----------MLDRCPELRHTGVVKIQPEFS---------AAHSAA--L
MLBr_00592|M.leprae_Br4923 --MLNVHRGYPSLDRSPELRHTGVVKIRSELD---------AATT-----
Mflv_3678|M.gilvum_PYR-GCK ------------------MRHTGVVKFRSQSS---------EAPTSAPLH
Mvan_2732|M.vanbaalenii_PYR-1 ------------------------------------------------MS
MAB_2750c|M.abscessus_ATCC_199 ------------------MKFGPGTSAGTAAT---------AAPSLNAAP
MSMEG_3121|M.smegmatis_MC2_155 ------------MEPSDEFRHNGVVKLRGGATSPVATGPSVPAADEVPGV
TH_1094|M.thermoresistible__bu --------------------------------------------------
Mb1495|M.bovis_AF2122/97 AAVSDGH--TRRAIVRLLLESGSITAGEIGDRLGLSAAGVRRHLDALIEA
Rv1460|M.tuberculosis_H37Rv AAVSDGH--TRRAIVRLLLESGSITAGEIGDRLGLSAAGVRRHLDALIEA
MAV_3319|M.avium_104 AAVPDGH--TRRAIVRLLLESGSITAGEIGDRLGLAAAGVRRHLDALIEA
MMAR_2265|M.marinum_M VAGSDGH--TRRAIVRLLLESGSITAGEIGDRLGLSAAGVRRHLDALIDA
MUL_1861|M.ulcerans_Agy99 VAGSDGH--TRRAIVRLLLESGSITAGEIGDRLGLSAAGVRRHLDALIDA
MLBr_00592|M.leprae_Br4923 VAVSDGH--TRRAVVRLLLESGSITAGEISDRLGLSAAGVRRHLDVLIEM
Mflv_3678|M.gilvum_PYR-GCK PVDGDHHGDTRSAIVRLLLESGPVTAGHIGQQLGLSAAGVRRHLDALIEA
Mvan_2732|M.vanbaalenii_PYR-1 PQAADGH--TRGAIVRLLLESGPVTASRIGERLGLSAAGVRRHLDALIEA
MAB_2750c|M.abscessus_ATCC_199 AASTEGQ--TRAAVVRLLLESGPITAGEIGERLGLSAAGVRRHLDALIDA
MSMEG_3121|M.smegmatis_MC2_155 ISTADGQ--TRRAIIKLLLE-GPITAGQIGERLGISAAGVRRHLDALIEA
TH_1094|M.thermoresistible__bu --------------------------------------------------
Mb1495|M.bovis_AF2122/97 GDAEASAAAPWQQVGRGRPAKRYRLTAAGRAKLDHSYDDLASAAMRQLRE
Rv1460|M.tuberculosis_H37Rv GDAEASAAAPWQQVGRGRPAKRYRLTAAGRAKLDHSYDDLASAAMRQLRE
MAV_3319|M.avium_104 GDAESIAPAAWQHAGRGRPAKRYRLTSAGRAKLDHAYDDLAAAAMRQLRE
MMAR_2265|M.marinum_M GDAESAPAAPWQQAGRGRPAKRYRLTPGGRAKLEHSYDDLAAAAMRQLRE
MUL_1861|M.ulcerans_Agy99 GDAESAPAAPWQQVGRGRPAKRYRLTPGGRAKLEHSYDDLAAAAMRQLRE
MLBr_00592|M.leprae_Br4923 GDAESATAAPWQQVGRGRPAKHYRLTAVGRAKLGHAYDTLASAAMRQLRE
Mflv_3678|M.gilvum_PYR-GCK GDAQANAAAAWQQEGRGRPAKRYRLTAAGRAKLEHTYDDLASAAMRQLRE
Mvan_2732|M.vanbaalenii_PYR-1 GEAQANAAAAWQQEGRGRPAKRYQLTSAGRAKLEHTYDDLASAAMRQLRE
MAB_2750c|M.abscessus_ATCC_199 GDAVAHPAAAWQHHGRGRPAKRFQLTADGRAKLNHAYDDLASAAMRQLRE
MSMEG_3121|M.smegmatis_MC2_155 GDAQASAAAAWQHTGRGRPAKRYRLTAAGRAKLGHSYDDLAVAAIRQLRE
TH_1094|M.thermoresistible__bu -----------------VPSDTX----XGRS-------------------
*:. **:
Mb1495|M.bovis_AF2122/97 IGGEEAVRTFARRRIDAILADVAPADGP-DDAALEAAAERIATALSKAGY
Rv1460|M.tuberculosis_H37Rv IGGEEAVRTFARRRIDAILADVAPADGP-DDAALEAAAERIATALSKAGY
MAV_3319|M.avium_104 IGGEEAVQAFARQRIDAILAGVPGAASA-ADDDIEAAAQRIATALTKAGY
MMAR_2265|M.marinum_M IGGDDAVRTFARRRIDAILAGVPAAASP-DDAAVEAAAERVASALTKAGY
MUL_1861|M.ulcerans_Agy99 IGGDDAVRTFARRRIDAILAGVPAAASP-DDAAVEAAAERVASALTKAGY
MLBr_00592|M.leprae_Br4923 IGGEDAVRTFARQRIDAILADVEPAGG---GADVEAAANRVASALSDAGY
Mflv_3678|M.gilvum_PYR-GCK IGGDDAVRTFARRRIDTILAGVEPVDDASRGEEIEAAADRIADALTGAGY
Mvan_2732|M.vanbaalenii_PYR-1 IGGEEAVRTFARRRIDTILSGVEHVD---GGEGVEAAAGQIAEALSAAGF
MAB_2750c|M.abscessus_ATCC_199 IGGDEAVRTFARRRIDTILGDLTIGAPQ---DDLPAAVGQVADALTRAGY
MSMEG_3121|M.smegmatis_MC2_155 IGGKDAVRTFARRRIDTILSGVTAGSG-----DVAQTAERVADAMTKAGY
TH_1094|M.thermoresistible__bu ----------PRRRIDAILAGVLGVSEG--AGDVESTAERVARALTRAGY
.*:***:**..: : :. ::* *:: **:
Mb1495|M.bovis_AF2122/97 VATTTRVGGP---IHGVQICQHHCPVSHVAEEFPELCETEQQAMAEVLGT
Rv1460|M.tuberculosis_H37Rv VATTTRVGGP---IHGVQICQHHCPVSHVAEEFPELCETEQQAMAEVLGT
MAV_3319|M.avium_104 VATTTQVGGP---IHGVQICQHHCPVSHVAEEFPELCDAEQQAMAEVLGT
MMAR_2265|M.marinum_M VATTTRVGGP---IHGVQICQHHCPVSHVAEEFPELCEAEQQAIAEVLGT
MUL_1861|M.ulcerans_Agy99 VATTTRVGGP---IHGVQICQHHCPVSHVAEEFPELCEAEQQAIAEVLGT
MLBr_00592|M.leprae_Br4923 VATTARVGGP---IHGVEICQHHCPVSHVAEEFPELCETEQQAIAEVLGT
Mflv_3678|M.gilvum_PYR-GCK ATTTPRVRGP---LSGVQICQHHCPVSHVAEQFPELCEAEQQAMSDVLGT
Mvan_2732|M.vanbaalenii_PYR-1 ATTTARVGGP---VGGVQICQHHCPVSHVAEEFPELCEAEQQAMAEVLGT
MAB_2750c|M.abscessus_ATCC_199 AASTQKVGGSGGFLQGVQICQHHCPVSHVAAEFPELCEAEQQAFAAVLGT
MSMEG_3121|M.smegmatis_MC2_155 ATTTAPVSGP---IRGVQICQHHCPVSHVAEEFPELCETEREAFAEILGT
TH_1094|M.thermoresistible__bu AATTSRVPGP---VDGVQICQHHCPVSHVAEEFPELCDAEREAFAEILDT
.::* * *. : **:************ :*****::*::*:: :*.*
Mb1495|M.bovis_AF2122/97 HVQRLATIVNGDCACTTHVPLSP---------------APSPRPPATSTE
Rv1460|M.tuberculosis_H37Rv HVQRLATIVNGDCACTTHVPLSP---------------APSPRPPATSTE
MAV_3319|M.avium_104 HVQRLATIVNGDCACTTHVPLAQTA------------KAPSPRRHTTSNK
MMAR_2265|M.marinum_M HVQRLATIVNGDCACTTHVPLTP---------------APSPRQHLTSTG
MUL_1861|M.ulcerans_Agy99 HVQRLATIVNGDCACTTHVPLTP---------------APSPRQHLTSTG
MLBr_00592|M.leprae_Br4923 HVQRLAMIVNGDCACTTHVPLMS---------------VPSKT-------
Mflv_3678|M.gilvum_PYR-GCK HVQRLATIVNGDCACTTHVPLVP------------------ER-------
Mvan_2732|M.vanbaalenii_PYR-1 HVQRLATIANGDFACTTHVPLSP------------------AGR------
MAB_2750c|M.abscessus_ATCC_199 HVQRLATIANGDCACTTHVPMQT-----------------SPPA------
MSMEG_3121|M.smegmatis_MC2_155 HVQRLATIVNGDCACTTHVPLDLDPPGEHTDHPKDVRSTARDRTTTPSDQ
TH_1094|M.thermoresistible__bu HVQRLATIVNGDNACTTHVPLPNRMPSHRKQPRKP--NRVQPRTATTSPQ
****** *.*** *******:
Mb1495|M.bovis_AF2122/97 GVSR
Rv1460|M.tuberculosis_H37Rv GASR
MAV_3319|M.avium_104 GASL
MMAR_2265|M.marinum_M GASA
MUL_1861|M.ulcerans_Agy99 GASA
MLBr_00592|M.leprae_Br4923 ----
Mflv_3678|M.gilvum_PYR-GCK ----
Mvan_2732|M.vanbaalenii_PYR-1 ----
MAB_2750c|M.abscessus_ATCC_199 ----
MSMEG_3121|M.smegmatis_MC2_155 GAST
TH_1094|M.thermoresistible__bu GASS