For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSISVEAAMRLAIDQAEQVKGATYPNPPVGAVILDRDGQVAGVGGTQPTGGPHAEVMALRAAAERAEGGT AVVTLEPCNHHGRTPPCVDGLVAAGISRVVYAVADPNPVAAGGSARMASSGIEVTSGVLSDEVAGGPLRE WLHKQRTGLPHVTWKFATSVDGRSAAADGSSQWITSAAARADVHRRRAAADAIVVGTGTVFVDDPSLTAR LPDGTLADRQPLRVVVGRREVSSDANVLNDDSRTMVIRTHDPHEVLRALSDRTDIMLEGGPTLAGAFLRA GVVNRILAYVAPILLGGPITAVDDVGVPSIAHAQRWRFDCVQTIGPDVLMSLVPN
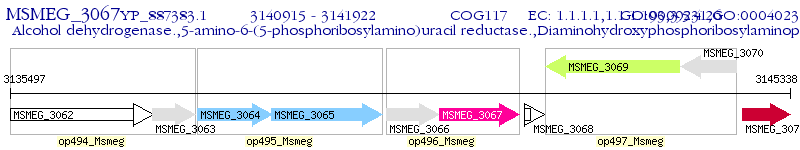
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3067 | ribD | - | 100% (335) | riboflavin biosynthesis protein RibD |
| M. smegmatis MC2 155 | MSMEG_6327 | - | 4e-11 | 33.96% (159) | cytidine and deoxycytidylate deaminase family protein |
| M. smegmatis MC2 155 | MSMEG_5219 | - | 7e-05 | 25.93% (162) | riboflavin biosynthesis protein RibD domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1444 | ribG | 1e-140 | 70.00% (330) | bifunctional diaminohydroxyphosphoribosylaminopyrimidine |
| M. gilvum PYR-GCK | Mflv_3727 | - | 1e-155 | 77.78% (333) | riboflavin biosynthesis protein RibD |
| M. tuberculosis H37Rv | Rv1409 | ribG | 1e-139 | 69.70% (330) | bifunctional diaminohydroxyphosphoribosylaminopyrimidine |
| M. leprae Br4923 | MLBr_00555 | ribG | 1e-141 | 71.52% (330) | putative bifunctional riboflavin-specific |
| M. abscessus ATCC 19977 | MAB_2808c | - | 1e-140 | 73.19% (332) | bifunctional riboflavin biosynthesis protein RibG |
| M. marinum M | MMAR_2218 | ribG | 1e-143 | 72.73% (330) | bifunctional riboflavin biosynthesis protein RibG |
| M. avium 104 | MAV_3368 | ribD | 1e-151 | 76.06% (330) | riboflavin biosynthesis protein RibD |
| M. thermoresistible (build 8) | TH_1007 | ribG | 1e-97 | 81.16% (207) | PROBABLE BIFUNCTIONAL riboflavin biosynthesis protein RIBG : |
| M. ulcerans Agy99 | MUL_1805 | ribG | 1e-143 | 72.73% (330) | bifunctional riboflavin biosynthesis protein RibG |
| M. vanbaalenii PYR-1 | Mvan_2690 | - | 1e-157 | 79.52% (332) | riboflavin biosynthesis protein RibD |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1444|M.bovis_AF2122/97 MNVEQVKSIDEAMGLAIEHSYQVKGTTYPNPPVGAVIVDPNGRIVGAGGT
Rv1409|M.tuberculosis_H37Rv MNVEQVKSIDEAMGLAIEHSYQVKGTTYPKPPVGAVIVDPNGRIVGAGGT
MMAR_2218|M.marinum_M MTVEQVKSLEEAMRLAIEQSMLVKGTTYPNPPVGAVIVDQEGRVVGIGGT
MUL_1805|M.ulcerans_Agy99 MTVEQVKSLEEAMRLAIEQSMLVKGTTYPNPPVGAVIVDQEGWVVGIGGT
MLBr_00555|M.leprae_Br4923 MNVQTVDSLDAAMRLAVEQSYLVKGATYPNPPVGAVIVDPDGRVVGVGAT
MAV_3368|M.avium_104 MTASLGDRLDAAMRLAIEQSNQVKGNTYPNPPVGAVVLDGRGEVVGVGGT
TH_1007|M.thermoresistible__bu --------------------------------------------------
Mflv_3727|M.gilvum_PYR-GCK ------MNLDAAMAAAIVQSERVKGRTYPNPPVGAVILDSDGEIAGVGAT
Mvan_2690|M.vanbaalenii_PYR-1 ------MKLDAAMAAAIEQSERVKGRTYPNPPVGAVILDRDGEIAGVGAT
MSMEG_3067|M.smegmatis_MC2_155 ----MSISVEAAMRLAIDQAEQVKGATYPNPPVGAVILDRDGQVAGVGGT
MAB_2808c|M.abscessus_ATCC_199 --MTNPIGLDEAMDLAIRAAEEVKGSTYPNPPVGAVILDAAGQVAGVGGT
Mb1444|M.bovis_AF2122/97 EPAGGDHAEVVALRRAGGLATGAIVVVTMEPCNHYGKTPPCVNALIEARV
Rv1409|M.tuberculosis_H37Rv EPAGGDHAEVVALRRAGGLAAGAIVVVTMEPCNHYGKTPPCVNALIEARV
MMAR_2218|M.marinum_M QPSGGDHAEVVALRKAGGLAAGAIAVVTMEPCNHFGKTPPCVNALIEARV
MUL_1805|M.ulcerans_Agy99 QPSGGDHAEVVALRKAGGLAAGAIAVVTMEPCNHFGKTPPCVNALIEARV
MLBr_00555|M.leprae_Br4923 EPAGGDHAEVVALRSAGVTAAGAIAVVTLEPCNHYGKTPPCVNALLEAKV
MAV_3368|M.avium_104 EPAGGDHAEILALRRAGDLAAGGTVVVTLEPCNHHGKTPPCVDALLEAGV
TH_1007|M.thermoresistible__bu --------------------------------------------------
Mflv_3727|M.gilvum_PYR-GCK APTGGPHAEVVALRRAGERAAGGTAVVTLEPCNHFGRTPPCVDALIDAGV
Mvan_2690|M.vanbaalenii_PYR-1 APPGGPHAEVVALRRAGERAVGGTAVVTLEPCNHFGRTPPCVDALLDAGI
MSMEG_3067|M.smegmatis_MC2_155 QPTGGPHAEVMALRAAAERAEGGTAVVTLEPCNHHGRTPPCVDGLVAAGI
MAB_2808c|M.abscessus_ATCC_199 RPPGQEHAEVVALAGAGAAARGGTAVVTLEPCNHQGRTGPCVDALAAAGV
Mb1444|M.bovis_AF2122/97 GTVVYAVADPNGIAGGGAGRLSAAGLQVRSGVLAEQVAAGPLREWLHKQR
Rv1409|M.tuberculosis_H37Rv GTVVYAVADPNGIAGGGAGRLSAAGLQVRSGVLAEQVAAGPLREWLHKQR
MMAR_2218|M.marinum_M GTVIYGVADPNGIAGGGAGRLSAAGLQVRSGVLADHVAAGPLREWLHKQR
MUL_1805|M.ulcerans_Agy99 GTVIYGVADPNGIAGGGAGRLSAAGLQVRSGVLADHVAAGPLREWLHKQR
MLBr_00555|M.leprae_Br4923 DKVAYAIADPNASAGGGAARLTAAGVHVQSGVLADLVTAGPLREWLHKQR
MAV_3368|M.avium_104 STVVYAVADPNPQAAGGAGRLQEAGVTVRSGLLADEVSGGPLREWLHKQR
TH_1007|M.thermoresistible__bu -------------------------------VLADEVAAGPLREWLHKTR
Mflv_3727|M.gilvum_PYR-GCK AAVAFAVADPNPAAAGGAARLAAAGVRVSAGVVADAVTGGPLREWLHKQR
Mvan_2690|M.vanbaalenii_PYR-1 AEVAFAVADPDPVAAGGAARLAESGVRVSAGVLAADVTGGPLREWLHKQR
MSMEG_3067|M.smegmatis_MC2_155 SRVVYAVADPNPVAAGGSARMASSGIEVTSGVLSDEVAGGPLREWLHKQR
MAB_2808c|M.abscessus_ATCC_199 SRVVYAVADPNPQAAGGAARLRTLGIEVLDGLASDRVSAGPLREWLHKQR
: : *:.********* *
Mb1444|M.bovis_AF2122/97 TGLPHVTWKYATSIDGRSAAADGSSQWISSEAARLDLHRRRAIADAILVG
Rv1409|M.tuberculosis_H37Rv TGLPHVTWKYATSIDGRSAAADGSSQWISSEAARLDLHRRRAIADAILVG
MMAR_2218|M.marinum_M TGLPHVTWKYATSIDGRSAAADGSSQWISSEAARADLHRRRAISDAIVVG
MUL_1805|M.ulcerans_Agy99 TGLPHVTWKYATSIDGRSAAADGSSQWISSEAARADLHRRRAISDAIVVG
MLBr_00555|M.leprae_Br4923 TGLPHVTWKYASSIDGRSAAADGSSRWLSSEASRADLHRRRATADAIVVG
MAV_3368|M.avium_104 TGLPHLTWKYASSVDGRSAAADGSSRWISSEASRLDLHRRRAAADAIVVG
TH_1007|M.thermoresistible__bu TGRPHVTWKFAASVDGRSAAADGTSQWITSEEARADVHRRRAAADAIVVG
Mflv_3727|M.gilvum_PYR-GCK TGKPHVTWKFAASVDGRSAAADGTSQWITSEASRADVHRRRAAADAIVAG
Mvan_2690|M.vanbaalenii_PYR-1 TGMPHVTWKFATSVDGRSAAADGSSQWITSEAARADVHRRRATADAIIVG
MSMEG_3067|M.smegmatis_MC2_155 TGLPHVTWKFATSVDGRSAAADGSSQWITSAAARADVHRRRAAADAIVVG
MAB_2808c|M.abscessus_ATCC_199 TGRPHVTWKYGASVDGRSAANDGTSQWITSEQSRADVHRRRAFADAVLVG
** **:***:.:*:****** **:*:*::* :* *:***** :**::.*
Mb1444|M.bovis_AF2122/97 TGTVLADDPALTARLADGSLAPQQPLRVVVGKRDIPPEARVLNDEARTMM
Rv1409|M.tuberculosis_H37Rv TGTVLADDPALTARLADGSLAPQQPLRVVVGKRDIPPEARVLNDEARTMM
MMAR_2218|M.marinum_M TGTVLVDDPTLTARLSDGSLAAEQPLRVVVGKRDIPPEAKVLNDDARTMM
MUL_1805|M.ulcerans_Agy99 TGTVLVDDPTLTARLSDGSLAAEQPLRVVVGKRDIPPEAKVLNDDARTMM
MLBr_00555|M.leprae_Br4923 TGTVLADNPTLTARLANGALADRQPLRVVVGMREIPLEANVLNDDSPTMV
MAV_3368|M.avium_104 TGTVLADDPALTARLPDGTLADRQPLRVVVGMREIPSEAKVLNDDSRTMV
TH_1007|M.thermoresistible__bu TGTVLADDPTLTARRPDGSLAERQPLRVVVGTRDISADANVLNDDSRTMV
Mflv_3727|M.gilvum_PYR-GCK TGTVFADDPALTARLPDGSLADHQPLRVVVGMREISPDATVLNDDSRTMV
Mvan_2690|M.vanbaalenii_PYR-1 TGTVFVDDPALTARLPDGSLADHQPLRVVVGKREISPDATVLNDDSRTMV
MSMEG_3067|M.smegmatis_MC2_155 TGTVFVDDPSLTARLPDGTLADRQPLRVVVGRREVSSDANVLNDDSRTMV
MAB_2808c|M.abscessus_ATCC_199 TGTVFADDPQLTARTPDNELVPHQPLRVVVGMREVSSDAKVLNDDSHTMV
****:.*:* **** .:. *. .******** *::. :* ****:: **:
Mb1444|M.bovis_AF2122/97 IRTHEPMEVLRALSDRTDVLLEGGPTLAGAFLRAGAINRILAYVAPILLG
Rv1409|M.tuberculosis_H37Rv IRTHEPMEVLRALSDRTDVLLEGGPTLAGAFLRAGAINRILAYVAPILLG
MMAR_2218|M.marinum_M IKTHEPVEVLRALSDRTDVLLEGGPTLAGAFLRAGAINRILAYVAPILLG
MUL_1805|M.ulcerans_Agy99 IKTHEPVEVLRALSDRTDVLLEGGPTLAGAFLRAGAINRILAYVAPILLG
MLBr_00555|M.leprae_Br4923 IRTHDPMEVLKALADRTDVLLEGGPTLAGAFLRAGTINRILAYVAPILLG
MAV_3368|M.avium_104 IRTHDPAEVLKAVSDRTDVLLEGGPTLAGAFVRAGLVNRILVYLAPTLLG
TH_1007|M.thermoresistible__bu IRTHDPHEVVRALADRTDVLLEGGPTLAGAFVRAGLVDRILAYVAPVLLG
Mflv_3727|M.gilvum_PYR-GCK IRTRDPHEVIQALTDRTDVLVEGGPTLAGAFLRAGAIDRILAYVAPILLG
Mvan_2690|M.vanbaalenii_PYR-1 IRTHNPHEVIKALADRTDVFVEGGPTLAGAFLRAGVIDRILAYVAPILLG
MSMEG_3067|M.smegmatis_MC2_155 IRTHDPHEVLRALSDRTDIMLEGGPTLAGAFLRAGVVNRILAYVAPILLG
MAB_2808c|M.abscessus_ATCC_199 IRTHDPAEVLAALGDRTDIILEGGPTLAGAFLRAGLVDRILAYLAPMLLG
*:*::* **: *: ****:::**********:*** ::***.*:** ***
Mb1444|M.bovis_AF2122/97 GPVTAVDDVGVSNITNALRWQFDSVEKVGPDLLLSLVAR---
Rv1409|M.tuberculosis_H37Rv GPVTAVDDVGVSNITNALRWQFDSVEKVGPDLLLSLVAR---
MMAR_2218|M.marinum_M GPVTAVDDVGVSSIAHALRWQFDGVERCGPDLLLSLVSR---
MUL_1805|M.ulcerans_Agy99 GPVTAVDDVGVSSIAHALRWQFDGVERCGPDLLLSLVSR---
MLBr_00555|M.leprae_Br4923 GPVTAVDDVGVPNIARALRWQFDGIDRAGSDLVISLVTR---
MAV_3368|M.avium_104 GPVTAVDDVGVPTIAKALRWQFDGVDRVGPDLLLSLVPRSG-
TH_1007|M.thermoresistible__bu GPVTVVDDVGVTTIGNAHRWRFDGMTSVGPDVLLSLVPR---
Mflv_3727|M.gilvum_PYR-GCK GPVTAVDDVGVSSITQAHRWRFDGMQAIGPDVLLSLVPN---
Mvan_2690|M.vanbaalenii_PYR-1 GPITAVDDVGVLSIAQAQRWRFDGMTAVGPDVLLSLTPV---
MSMEG_3067|M.smegmatis_MC2_155 GPITAVDDVGVPSIAHAQRWRFDCVQTIGPDVLMSLVPN---
MAB_2808c|M.abscessus_ATCC_199 GNVPAVEDVGVTTLSRALRWEYDQIERMGPDVLLSLVPRLPV
* :..*:**** .: .* **.:* : *.*:::**..